Gene
KWMTBOMO15218
Pre Gene Modal
BGIBMGA004959
Annotation
PREDICTED:_beta-1?3-galactosyltransferase_2_[Bombyx_mori]
Full name
Hexosyltransferase
Location in the cell
PlasmaMembrane Reliability : 1.595
Sequence
CDS
ATGGTGAAAAGAAAATATGTTTTAGCTTCCATTTTAGCCGTAACTTTGATTAGTTGGATATATTGGAGTTCAAACCATGAAGTAAATAACACTATAAAAATACTTTCTGAACAAGAAAGTTTTAAACATACAATTATTCAACCAAGTGACAAATTGTGTACAGGAAATCCCTTTCTGGTAATTATAGTTACATCGTATGTGGGCCATATTGAACTACGAAGTGCTCACAGAAGAGCCATGCCTCAAAAACTTCTGGAAGCCATGAACGCTAAAAGAGTATTCTTATTAGCTAAAATTCCTGAAAAAGACAAAATCATCACGCAAGAGGCAATAATAGACGAAAGCAATATATTCGGAGATATCCTGCAAGGATCTTTCGTGGAAAATTATAGAAATTTAACGTTCAAGCATCTGATGGGCTTGCAGTGGGCCTCCACAGCGTGCAGCAATGCATCGTACATTTTAAAAGTCGACGACGATACCGTTTTTAATTTAGAAAGGACTTATCGATTACTAGAAACTTTAGATGCAAATGACATTTTCCTTCTGGGTTACATTCTCAATAATACGAAGCCGAGAAGGAATAAAGAAAACAAATGGTATGTTACTCGGGAAGAATACTCTAGAAAAAACTACCCTGCTTATTTATCGGGGTGGTATTACATAACAACGCCAAAAACGGCCCGCAAAATAACCGAGGAAGCTATATACCATCCTAAATTCTGGATTGACGACATTTTAATTACCGGTATGCTCACTGAAGCTTTACATATTAAATTACGACAATTGCCAAGAGATTATTGGTTGGAATATTATGAGTTACTCGAGTGTTGTTTAAGAGATATGATAAATAAACAAATTAATTGCGATTCGGTAGTTGGACCGAACGGTGGGAGAGAGAATTTGATTGTAGAATTTAATGACGCAATAAGAAACTGTGATAAATCGAAAAAGTGCACGAAACGTGATGAAAGCACGCCGTTAAAGAAAGTATGCATTGTGAACAGTGAAAGAACTATATTTAGTAATGGTCAACCTCAAGTGGAGTTATATAAATTATAA
Protein
MVKRKYVLASILAVTLISWIYWSSNHEVNNTIKILSEQESFKHTIIQPSDKLCTGNPFLVIIVTSYVGHIELRSAHRRAMPQKLLEAMNAKRVFLLAKIPEKDKIITQEAIIDESNIFGDILQGSFVENYRNLTFKHLMGLQWASTACSNASYILKVDDDTVFNLERTYRLLETLDANDIFLLGYILNNTKPRRNKENKWYVTREEYSRKNYPAYLSGWYYITTPKTARKITEEAIYHPKFWIDDILITGMLTEALHIKLRQLPRDYWLEYYELLECCLRDMINKQINCDSVVGPNGGRENLIVEFNDAIRNCDKSKKCTKRDESTPLKKVCIVNSERTIFSNGQPQVELYKL
Summary
Similarity
Belongs to the glycosyltransferase 31 family.
Feature
chain Hexosyltransferase
Uniprot
A0A2A4K3H2
A0A2H1VLQ7
A0A212FIN0
A0A2A4K4G7
A0A194PG22
A0A194RK00
+ More
A0A2W1BIX3 W8BXP2 A0A1I8QA81 A0A336LNU1 A0A1Y1M0V1 A0A0A1WFV8 A0A0L0C2S6 A0A3B0JXC5 A0A0K8ULI2 B4NCY4 Q8IRZ0 B3MYN6 A0A1W4V8G8 Q95RP8 A0A1B0D975 Q6W4K1 A0A182IS38 Q6W4J1 Q6W4I1 Q9V394 A0A084VHC0 A0A0K8TKC0 A0A146LQ42 Q6W4H4 Q6W4H5 Q29JI0 A0A182FKS3 A0A0M5J5U7 Q6W4H6 B4H316 B4PWJ3 A0A182QP61 B4L5Z4 B0WRM9 W5J733 A0A1B0C8A3 U4U2N6 A0A182R6B9 B4I8Z8 B4JMU4 Q6W4H8 B3P945 A0A1B6CHV2 N6TYP8 B4R723 B4M7I8 D1ZZD3 A0A1A9W6C0 Q176W7 A0A182NCF9 A0A026WKN0 A0A182MAN7 A0A1W4W4G2 A0A182T2Y9 A0A182VZ22 A0A1I8MYY7 A0A182PRK0 A0A182XVY2 A0A182XBA6 A0A182TTR9 A0A182V6C4 Q7PYM6 A0A182I048 A0A182LH19 A0A182K2F9 A0A158P1V3 A0A2M4CN80 K7J540 A0A151K2G4 A0A232F653 A0A195AVS5 A0A1B6LE77 A0A151WNZ4 X2JD78 A0A2R7VV42 E9J3S7 F4W626 A0A067RJ13 A0A195DMR8 A0A195ERA0 T1IAH9 E2AGN7 A0A023F0A9 E2BU61 A0A0V0GAB1 A0A1B6K090 A0A088AW17 A0A2A3E734 A0A2P8Z7G1
A0A2W1BIX3 W8BXP2 A0A1I8QA81 A0A336LNU1 A0A1Y1M0V1 A0A0A1WFV8 A0A0L0C2S6 A0A3B0JXC5 A0A0K8ULI2 B4NCY4 Q8IRZ0 B3MYN6 A0A1W4V8G8 Q95RP8 A0A1B0D975 Q6W4K1 A0A182IS38 Q6W4J1 Q6W4I1 Q9V394 A0A084VHC0 A0A0K8TKC0 A0A146LQ42 Q6W4H4 Q6W4H5 Q29JI0 A0A182FKS3 A0A0M5J5U7 Q6W4H6 B4H316 B4PWJ3 A0A182QP61 B4L5Z4 B0WRM9 W5J733 A0A1B0C8A3 U4U2N6 A0A182R6B9 B4I8Z8 B4JMU4 Q6W4H8 B3P945 A0A1B6CHV2 N6TYP8 B4R723 B4M7I8 D1ZZD3 A0A1A9W6C0 Q176W7 A0A182NCF9 A0A026WKN0 A0A182MAN7 A0A1W4W4G2 A0A182T2Y9 A0A182VZ22 A0A1I8MYY7 A0A182PRK0 A0A182XVY2 A0A182XBA6 A0A182TTR9 A0A182V6C4 Q7PYM6 A0A182I048 A0A182LH19 A0A182K2F9 A0A158P1V3 A0A2M4CN80 K7J540 A0A151K2G4 A0A232F653 A0A195AVS5 A0A1B6LE77 A0A151WNZ4 X2JD78 A0A2R7VV42 E9J3S7 F4W626 A0A067RJ13 A0A195DMR8 A0A195ERA0 T1IAH9 E2AGN7 A0A023F0A9 E2BU61 A0A0V0GAB1 A0A1B6K090 A0A088AW17 A0A2A3E734 A0A2P8Z7G1
EC Number
2.4.1.-
Pubmed
22118469
26354079
28756777
24495485
28004739
25830018
+ More
26108605 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24438588 26369729 26823975 15632085 17550304 20920257 23761445 23537049 18362917 19820115 17510324 24508170 30249741 25315136 25244985 12364791 14747013 17210077 20966253 21347285 20075255 28648823 21282665 21719571 24845553 20798317 25474469 29403074
26108605 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24438588 26369729 26823975 15632085 17550304 20920257 23761445 23537049 18362917 19820115 17510324 24508170 30249741 25315136 25244985 12364791 14747013 17210077 20966253 21347285 20075255 28648823 21282665 21719571 24845553 20798317 25474469 29403074
EMBL
NWSH01000170
PCG78805.1
ODYU01003246
SOQ41748.1
AGBW02008360
OWR53586.1
+ More
PCG78804.1 KQ459604 KPI92336.1 KQ460045 KPJ18158.1 KZ150014 PZC75009.1 GAMC01002473 JAC04083.1 UFQT01000038 SSX18431.1 GEZM01043560 GEZM01043559 GEZM01043558 GEZM01043557 GEZM01043556 GEZM01043555 JAV79021.1 GBXI01016393 JAC97898.1 JRES01000981 KNC26537.1 OUUW01000011 SPP86727.1 GDHF01024757 JAI27557.1 CH964239 EDW82693.1 AE014298 BT125836 AAN09009.1 ADR83721.1 CH902632 EDV32730.1 AY061226 AAF45486.2 AAL28774.1 AJVK01004269 AY312642 AY312643 AY312644 AY312645 AY312646 AY312647 AY312648 AY312650 AY312651 AY312652 AAQ67456.1 AAQ67457.1 AAQ67458.1 AAQ67459.1 AAQ67460.1 AAQ67461.1 AAQ67462.1 AAQ67464.1 AAQ67465.1 AAQ67466.1 AY312649 AY312653 AY312654 AY312655 AY312656 AY312657 AY312658 AY312659 AY312660 AY312661 AAQ67463.1 AAQ67467.1 AAQ67468.1 AAQ67469.1 AAQ67470.1 AAQ67471.1 AAQ67472.1 AAQ67473.1 AAQ67474.1 AAQ67475.1 AY312662 AY312663 AY312664 AY312665 AAQ67476.1 AAQ67477.1 AAQ67478.1 AAQ67479.1 AL050231 CAB65849.1 ATLV01013151 KE524842 KFB37364.1 GDAI01002831 JAI14772.1 GDHC01014573 GDHC01009777 JAQ04056.1 JAQ08852.1 AY312670 AAQ67484.1 AY312669 AAQ67483.1 CH379063 EAL32321.1 CP012528 ALC48406.1 AY312667 AY312668 AAQ67481.1 AAQ67482.1 CH479205 EDW30709.1 CM000162 EDX00762.1 AXCN02000019 CH933811 EDW06603.1 DS232057 EDS33429.1 ADMH02002044 ETN59786.1 AJWK01000296 KB632031 ERL88154.1 CH480825 EDW43679.1 CH916371 EDV92037.1 AY312666 AAQ67480.1 CH954183 EDV45341.1 GEDC01024327 GEDC01017692 JAS12971.1 JAS19606.1 APGK01046660 KB741064 ENN74435.1 CM000366 EDX16734.1 CH940653 EDW62755.2 KQ971338 EFA02353.1 CH477381 EAT42205.1 KK107163 QOIP01000002 EZA56538.1 RLU25709.1 AXCM01004517 AAAB01008987 EAA01640.3 APCN01002043 ADTU01006889 GGFL01002608 MBW66786.1 AAZX01000348 LKEX01012869 KYN50203.1 NNAY01000936 OXU25837.1 KQ976731 KYM76343.1 GEBQ01017998 JAT21979.1 KQ982892 KYQ49609.1 AHN59203.1 KK854029 PTY09795.1 GL768069 EFZ12456.1 GL887707 EGI70161.1 KK852478 KDR23013.1 KQ980724 KYN14168.1 KQ982021 KYN30432.1 ACPB03010649 GL439357 EFN67402.1 GBBI01004478 JAC14234.1 GL450581 EFN80778.1 GECL01001224 JAP04900.1 GECU01002842 JAT04865.1 KZ288360 PBC27036.1 PYGN01000162 PSN52426.1
PCG78804.1 KQ459604 KPI92336.1 KQ460045 KPJ18158.1 KZ150014 PZC75009.1 GAMC01002473 JAC04083.1 UFQT01000038 SSX18431.1 GEZM01043560 GEZM01043559 GEZM01043558 GEZM01043557 GEZM01043556 GEZM01043555 JAV79021.1 GBXI01016393 JAC97898.1 JRES01000981 KNC26537.1 OUUW01000011 SPP86727.1 GDHF01024757 JAI27557.1 CH964239 EDW82693.1 AE014298 BT125836 AAN09009.1 ADR83721.1 CH902632 EDV32730.1 AY061226 AAF45486.2 AAL28774.1 AJVK01004269 AY312642 AY312643 AY312644 AY312645 AY312646 AY312647 AY312648 AY312650 AY312651 AY312652 AAQ67456.1 AAQ67457.1 AAQ67458.1 AAQ67459.1 AAQ67460.1 AAQ67461.1 AAQ67462.1 AAQ67464.1 AAQ67465.1 AAQ67466.1 AY312649 AY312653 AY312654 AY312655 AY312656 AY312657 AY312658 AY312659 AY312660 AY312661 AAQ67463.1 AAQ67467.1 AAQ67468.1 AAQ67469.1 AAQ67470.1 AAQ67471.1 AAQ67472.1 AAQ67473.1 AAQ67474.1 AAQ67475.1 AY312662 AY312663 AY312664 AY312665 AAQ67476.1 AAQ67477.1 AAQ67478.1 AAQ67479.1 AL050231 CAB65849.1 ATLV01013151 KE524842 KFB37364.1 GDAI01002831 JAI14772.1 GDHC01014573 GDHC01009777 JAQ04056.1 JAQ08852.1 AY312670 AAQ67484.1 AY312669 AAQ67483.1 CH379063 EAL32321.1 CP012528 ALC48406.1 AY312667 AY312668 AAQ67481.1 AAQ67482.1 CH479205 EDW30709.1 CM000162 EDX00762.1 AXCN02000019 CH933811 EDW06603.1 DS232057 EDS33429.1 ADMH02002044 ETN59786.1 AJWK01000296 KB632031 ERL88154.1 CH480825 EDW43679.1 CH916371 EDV92037.1 AY312666 AAQ67480.1 CH954183 EDV45341.1 GEDC01024327 GEDC01017692 JAS12971.1 JAS19606.1 APGK01046660 KB741064 ENN74435.1 CM000366 EDX16734.1 CH940653 EDW62755.2 KQ971338 EFA02353.1 CH477381 EAT42205.1 KK107163 QOIP01000002 EZA56538.1 RLU25709.1 AXCM01004517 AAAB01008987 EAA01640.3 APCN01002043 ADTU01006889 GGFL01002608 MBW66786.1 AAZX01000348 LKEX01012869 KYN50203.1 NNAY01000936 OXU25837.1 KQ976731 KYM76343.1 GEBQ01017998 JAT21979.1 KQ982892 KYQ49609.1 AHN59203.1 KK854029 PTY09795.1 GL768069 EFZ12456.1 GL887707 EGI70161.1 KK852478 KDR23013.1 KQ980724 KYN14168.1 KQ982021 KYN30432.1 ACPB03010649 GL439357 EFN67402.1 GBBI01004478 JAC14234.1 GL450581 EFN80778.1 GECL01001224 JAP04900.1 GECU01002842 JAT04865.1 KZ288360 PBC27036.1 PYGN01000162 PSN52426.1
Proteomes
UP000218220
UP000007151
UP000053268
UP000053240
UP000095300
UP000037069
+ More
UP000268350 UP000007798 UP000000803 UP000007801 UP000192221 UP000092462 UP000075880 UP000030765 UP000001819 UP000069272 UP000092553 UP000008744 UP000002282 UP000075886 UP000009192 UP000002320 UP000000673 UP000092461 UP000030742 UP000075900 UP000001292 UP000001070 UP000008711 UP000019118 UP000000304 UP000008792 UP000007266 UP000091820 UP000008820 UP000075884 UP000053097 UP000279307 UP000075883 UP000192223 UP000075901 UP000075920 UP000095301 UP000075885 UP000076408 UP000076407 UP000075902 UP000075903 UP000007062 UP000075840 UP000075882 UP000075881 UP000005205 UP000002358 UP000078542 UP000215335 UP000078540 UP000075809 UP000007755 UP000027135 UP000078492 UP000078541 UP000015103 UP000000311 UP000008237 UP000005203 UP000242457 UP000245037
UP000268350 UP000007798 UP000000803 UP000007801 UP000192221 UP000092462 UP000075880 UP000030765 UP000001819 UP000069272 UP000092553 UP000008744 UP000002282 UP000075886 UP000009192 UP000002320 UP000000673 UP000092461 UP000030742 UP000075900 UP000001292 UP000001070 UP000008711 UP000019118 UP000000304 UP000008792 UP000007266 UP000091820 UP000008820 UP000075884 UP000053097 UP000279307 UP000075883 UP000192223 UP000075901 UP000075920 UP000095301 UP000075885 UP000076408 UP000076407 UP000075902 UP000075903 UP000007062 UP000075840 UP000075882 UP000075881 UP000005205 UP000002358 UP000078542 UP000215335 UP000078540 UP000075809 UP000007755 UP000027135 UP000078492 UP000078541 UP000015103 UP000000311 UP000008237 UP000005203 UP000242457 UP000245037
Pfam
PF01762 Galactosyl_T
SUPFAM
SSF53448
SSF53448
ProteinModelPortal
A0A2A4K3H2
A0A2H1VLQ7
A0A212FIN0
A0A2A4K4G7
A0A194PG22
A0A194RK00
+ More
A0A2W1BIX3 W8BXP2 A0A1I8QA81 A0A336LNU1 A0A1Y1M0V1 A0A0A1WFV8 A0A0L0C2S6 A0A3B0JXC5 A0A0K8ULI2 B4NCY4 Q8IRZ0 B3MYN6 A0A1W4V8G8 Q95RP8 A0A1B0D975 Q6W4K1 A0A182IS38 Q6W4J1 Q6W4I1 Q9V394 A0A084VHC0 A0A0K8TKC0 A0A146LQ42 Q6W4H4 Q6W4H5 Q29JI0 A0A182FKS3 A0A0M5J5U7 Q6W4H6 B4H316 B4PWJ3 A0A182QP61 B4L5Z4 B0WRM9 W5J733 A0A1B0C8A3 U4U2N6 A0A182R6B9 B4I8Z8 B4JMU4 Q6W4H8 B3P945 A0A1B6CHV2 N6TYP8 B4R723 B4M7I8 D1ZZD3 A0A1A9W6C0 Q176W7 A0A182NCF9 A0A026WKN0 A0A182MAN7 A0A1W4W4G2 A0A182T2Y9 A0A182VZ22 A0A1I8MYY7 A0A182PRK0 A0A182XVY2 A0A182XBA6 A0A182TTR9 A0A182V6C4 Q7PYM6 A0A182I048 A0A182LH19 A0A182K2F9 A0A158P1V3 A0A2M4CN80 K7J540 A0A151K2G4 A0A232F653 A0A195AVS5 A0A1B6LE77 A0A151WNZ4 X2JD78 A0A2R7VV42 E9J3S7 F4W626 A0A067RJ13 A0A195DMR8 A0A195ERA0 T1IAH9 E2AGN7 A0A023F0A9 E2BU61 A0A0V0GAB1 A0A1B6K090 A0A088AW17 A0A2A3E734 A0A2P8Z7G1
A0A2W1BIX3 W8BXP2 A0A1I8QA81 A0A336LNU1 A0A1Y1M0V1 A0A0A1WFV8 A0A0L0C2S6 A0A3B0JXC5 A0A0K8ULI2 B4NCY4 Q8IRZ0 B3MYN6 A0A1W4V8G8 Q95RP8 A0A1B0D975 Q6W4K1 A0A182IS38 Q6W4J1 Q6W4I1 Q9V394 A0A084VHC0 A0A0K8TKC0 A0A146LQ42 Q6W4H4 Q6W4H5 Q29JI0 A0A182FKS3 A0A0M5J5U7 Q6W4H6 B4H316 B4PWJ3 A0A182QP61 B4L5Z4 B0WRM9 W5J733 A0A1B0C8A3 U4U2N6 A0A182R6B9 B4I8Z8 B4JMU4 Q6W4H8 B3P945 A0A1B6CHV2 N6TYP8 B4R723 B4M7I8 D1ZZD3 A0A1A9W6C0 Q176W7 A0A182NCF9 A0A026WKN0 A0A182MAN7 A0A1W4W4G2 A0A182T2Y9 A0A182VZ22 A0A1I8MYY7 A0A182PRK0 A0A182XVY2 A0A182XBA6 A0A182TTR9 A0A182V6C4 Q7PYM6 A0A182I048 A0A182LH19 A0A182K2F9 A0A158P1V3 A0A2M4CN80 K7J540 A0A151K2G4 A0A232F653 A0A195AVS5 A0A1B6LE77 A0A151WNZ4 X2JD78 A0A2R7VV42 E9J3S7 F4W626 A0A067RJ13 A0A195DMR8 A0A195ERA0 T1IAH9 E2AGN7 A0A023F0A9 E2BU61 A0A0V0GAB1 A0A1B6K090 A0A088AW17 A0A2A3E734 A0A2P8Z7G1
Ontologies
GO
PANTHER
Topology
Subcellular location
Golgi apparatus membrane
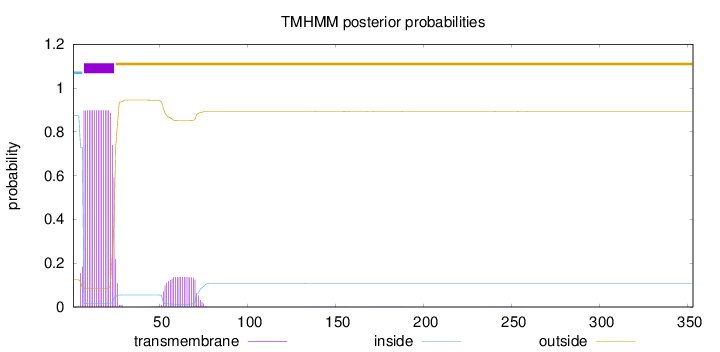
Length:
353
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
19.02459
Exp number, first 60 AAs:
17.46126
Total prob of N-in:
0.87490
POSSIBLE N-term signal
sequence
inside
1 - 6
TMhelix
7 - 24
outside
25 - 353
Population Genetic Test Statistics
Pi
162.691903
Theta
130.165026
Tajima's D
0.784912
CLR
0.125853
CSRT
0.6007199640018
Interpretation
Uncertain
