Pre Gene Modal
BGIBMGA004962
Annotation
PREDICTED:_intron-binding_protein_aquarius_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 2.181
Sequence
CDS
ATGAAGACTGAAACCCCACAGCCTACGGATAAACTTAAAATACCATTACCGACACTATCACAGATTAACGCGGATAGAATAACACAGCTTGCAAATCAGTACTGGTCACCGCAAACCAAGGAATCTCACTTACCTTATGATGCTTCTATTGTGGAGTCAATATACCATGCAGAAATCCAGGGTTCAAGTTTCTCAGTTCGACGCATCATGATGTTAGAGTTCTCACAATACCTCGAGAACTATCTGTGGCCATACTATGAAGCTGGCAAGGCATCTCACTCACACATGATGTCTATAATAGTGATGATAAATGAGAAGTTCCGTGAGCGAGTTCCCGCTTGGCAGGCATTTTTAAAGCAACCGGAGCACTTTCCGGCCTTCTTTGAACAAGTCCTTCGTGCCAGTATTGCTGAAGACGACACTAGTCGTAACATGAGGGATCAGACTGCATTACTGCTATTCCTGAATCATTGCTTCGGCAGCATGGAAGTAGAGCTCTGCAGGGATCAAGTCAAGAGGCTTGTGTCCCTTAGTATGTGGATTAGTTTACAAGAAGGCAGAAGAAATCAGGAATTCAAAGCGGTACCAAAGTGGCGTAAATATTGGAGAGCCATACAGAAAAAGGACAAGCCAGAGTTATTGGAAAAATTGAACTGGGAACGAAGATATTTGCAAAGACTGATGATCAAGTTTATGGGAATTCTAGAGAAAATACCCGAAAACGGGGAAATTGATTCGTATGTTGTGAGGTACTGCGAACGTTTCCTGGAATTGATGATAGACCTGGAGGCTTTACTGCCGACGCGCCGTTTCTTCAACACGGTGATGGACGACTGTCATCTCGTGGTGCGCTGTCAGATGTCAGCACTCGCCCAGAGACCAGAGGGGCAACTCTTTACACAAGTAAGTAATGCAATATTTAATTGA
Protein
MKTETPQPTDKLKIPLPTLSQINADRITQLANQYWSPQTKESHLPYDASIVESIYHAEIQGSSFSVRRIMMLEFSQYLENYLWPYYEAGKASHSHMMSIIVMINEKFRERVPAWQAFLKQPEHFPAFFEQVLRASIAEDDTSRNMRDQTALLLFLNHCFGSMEVELCRDQVKRLVSLSMWISLQEGRRNQEFKAVPKWRKYWRAIQKKDKPELLEKLNWERRYLQRLMIKFMGILEKIPENGEIDSYVVRYCERFLELMIDLEALLPTRRFFNTVMDDCHLVVRCQMSALAQRPEGQLFTQVSNAIFN
Summary
Similarity
Belongs to the CWF11 family.
Uniprot
H9J621
A0A2H1VG59
A0A2A4K449
A0A194PGA1
A0A212FIM0
A0A194RLH2
+ More
A0A2W1BIW6 A0A2J7PR16 A0A1B6MBW9 A0A1B6DEG6 A0A0M9A4E5 A0A0L7QME1 A0A067R9P9 E2AK71 A0A1B0D9S2 A0A1B6HYD6 A0A154PIH9 A0A1W4WW62 A0A1B0CCV8 A0A1B6ED22 A0A0J7NUM9 A0A1B0GNZ8 E9IMC0 A0A139WG03 E2BAV8 A0A1B6EMU7 A0A1B6GWX1 F4WFF0 A0A0C9QDX1 U4U6K8 N6SSY4 B0W921 A0A2P8ZK85 A0A182JI35 Q16PU5 W5JUG5 A0A2A3EDJ5 A0A087ZT71 A0A182HA09 A0A0C9QLK7 A0A182MNY7 A0A182FA40 A0A182RFK2 A0A084VTL6 E0VSM8 A0A182P426 A0A182YG14 A0A1Y1LXY0 E9GFT7 A0A3L8DKL5 A0A026WA84 A0A182XTK9 A0A182TXC2 A0A182VWZ2 A0A182KMM2 A0A182I873 A0A195CQN9 A0A2M4ATM6 A0A182USE3 A0A182NDY5 A0A182JSW3 K7IQG0 A0A158NCM0 A0A232EXP4 A0A1J1HTU8 A0A2M4CVB5 A0A0P6J137 A0A2M4A8F1 A0A2M4A8E8 A0A0P5W7X3 A0A0P5TQM9 A0A0P4YCV3 A0A0P6HES3 A0A0P5LI99 A0A0P5MV71 A0A151X614 A0A195F9R1 B4K9Z3 A0A0P4XLG9 A0A0T6BDX9 A0A195BXI1 A0A0P5LU86 A0A0P5A7Y0 A0A195EMU4 A0A0P6B471 A0A0P4Z9N0 A0A0P5UI14 A0A162C8N0 A0A0P5R972 A0A0P5P7U8 A0A0P5P7T0 A0A0P6JI64 A0A0P6DTT8 A0A0P5ZUZ1 B4LXY8 F5HM53 B4JG10 B4NK55 V5H2X1 A0A1W4WBZ9
A0A2W1BIW6 A0A2J7PR16 A0A1B6MBW9 A0A1B6DEG6 A0A0M9A4E5 A0A0L7QME1 A0A067R9P9 E2AK71 A0A1B0D9S2 A0A1B6HYD6 A0A154PIH9 A0A1W4WW62 A0A1B0CCV8 A0A1B6ED22 A0A0J7NUM9 A0A1B0GNZ8 E9IMC0 A0A139WG03 E2BAV8 A0A1B6EMU7 A0A1B6GWX1 F4WFF0 A0A0C9QDX1 U4U6K8 N6SSY4 B0W921 A0A2P8ZK85 A0A182JI35 Q16PU5 W5JUG5 A0A2A3EDJ5 A0A087ZT71 A0A182HA09 A0A0C9QLK7 A0A182MNY7 A0A182FA40 A0A182RFK2 A0A084VTL6 E0VSM8 A0A182P426 A0A182YG14 A0A1Y1LXY0 E9GFT7 A0A3L8DKL5 A0A026WA84 A0A182XTK9 A0A182TXC2 A0A182VWZ2 A0A182KMM2 A0A182I873 A0A195CQN9 A0A2M4ATM6 A0A182USE3 A0A182NDY5 A0A182JSW3 K7IQG0 A0A158NCM0 A0A232EXP4 A0A1J1HTU8 A0A2M4CVB5 A0A0P6J137 A0A2M4A8F1 A0A2M4A8E8 A0A0P5W7X3 A0A0P5TQM9 A0A0P4YCV3 A0A0P6HES3 A0A0P5LI99 A0A0P5MV71 A0A151X614 A0A195F9R1 B4K9Z3 A0A0P4XLG9 A0A0T6BDX9 A0A195BXI1 A0A0P5LU86 A0A0P5A7Y0 A0A195EMU4 A0A0P6B471 A0A0P4Z9N0 A0A0P5UI14 A0A162C8N0 A0A0P5R972 A0A0P5P7U8 A0A0P5P7T0 A0A0P6JI64 A0A0P6DTT8 A0A0P5ZUZ1 B4LXY8 F5HM53 B4JG10 B4NK55 V5H2X1 A0A1W4WBZ9
Pubmed
EMBL
BABH01029882
ODYU01002195
SOQ39372.1
NWSH01000170
PCG78796.1
KQ459604
+ More
KPI92327.1 AGBW02008360 OWR53577.1 KQ460045 KPJ18165.1 KZ150014 PZC74999.1 NEVH01022633 PNF18780.1 GEBQ01006545 JAT33432.1 GEDC01013229 JAS24069.1 KQ435736 KOX77020.1 KQ414894 KOC59701.1 KK852653 KDR19357.1 GL440194 EFN66175.1 AJVK01028429 GECU01028005 JAS79701.1 KQ434905 KZC11294.1 AJWK01007142 GEDC01001473 JAS35825.1 LBMM01001568 KMQ96075.1 AJVK01032962 GL764129 EFZ18454.1 KQ971351 KYB26826.1 GL446861 EFN87165.1 GECZ01030520 JAS39249.1 GECZ01002857 JAS66912.1 GL888115 EGI67201.1 GBYB01001519 JAG71286.1 KB631802 ERL86251.1 APGK01057806 KB741282 ENN70799.1 DS231861 EDS39459.1 PYGN01000032 PSN56916.1 CH477772 EAT36383.1 ADMH02000048 ETN67992.1 KZ288292 PBC29091.1 JXUM01030890 KQ560901 KXJ80422.1 GBYB01001520 GBYB01001521 GBYB01001522 GBYB01001523 GBYB01007951 GBYB01011553 JAG71287.1 JAG71288.1 JAG71289.1 JAG71290.1 JAG77718.1 JAG81320.1 AXCM01001491 ATLV01016361 KE525079 KFB41310.1 AAZO01005147 DS235751 EEB16384.1 GEZM01046474 JAV77210.1 GL732542 EFX81697.1 QOIP01000007 RLU20378.1 KK107364 EZA51939.1 APCN01002661 KQ977408 KYN02960.1 GGFK01010806 MBW44127.1 ADTU01011962 ADTU01011963 ADTU01011964 NNAY01001737 OXU23077.1 CVRI01000021 CRK91496.1 GGFL01004600 MBW68778.1 GDUN01000236 JAN95683.1 GGFK01003762 MBW37083.1 GGFK01003689 MBW37010.1 GDIP01090024 JAM13691.1 GDIP01125922 JAL77792.1 GDIP01231964 JAI91437.1 GDIQ01033471 JAN61266.1 GDIQ01169244 JAK82481.1 GDIQ01150751 JAL00975.1 KQ982490 KYQ55807.1 KQ981727 KYN36952.1 CH933806 EDW15641.2 GDIP01239663 JAI83738.1 LJIG01001402 KRT85538.1 KQ976396 KYM93005.1 GDIQ01165505 JAK86220.1 GDIP01207218 JAJ16184.1 KQ978691 KYN29222.1 GDIP01022404 JAM81311.1 GDIP01215707 JAJ07695.1 GDIP01114019 JAL89695.1 LRGB01001348 KZS12822.1 GDIQ01113114 JAL38612.1 GDIQ01132462 JAL19264.1 GDIQ01132463 JAL19263.1 GDIQ01007414 JAN87323.1 GDIQ01073034 JAN21703.1 GDIP01038685 JAM65030.1 CH940650 EDW66854.2 AAAB01008978 EGK97364.1 CH916369 EDV92549.1 CH964272 EDW85097.1 GALX01001278 JAB67188.1
KPI92327.1 AGBW02008360 OWR53577.1 KQ460045 KPJ18165.1 KZ150014 PZC74999.1 NEVH01022633 PNF18780.1 GEBQ01006545 JAT33432.1 GEDC01013229 JAS24069.1 KQ435736 KOX77020.1 KQ414894 KOC59701.1 KK852653 KDR19357.1 GL440194 EFN66175.1 AJVK01028429 GECU01028005 JAS79701.1 KQ434905 KZC11294.1 AJWK01007142 GEDC01001473 JAS35825.1 LBMM01001568 KMQ96075.1 AJVK01032962 GL764129 EFZ18454.1 KQ971351 KYB26826.1 GL446861 EFN87165.1 GECZ01030520 JAS39249.1 GECZ01002857 JAS66912.1 GL888115 EGI67201.1 GBYB01001519 JAG71286.1 KB631802 ERL86251.1 APGK01057806 KB741282 ENN70799.1 DS231861 EDS39459.1 PYGN01000032 PSN56916.1 CH477772 EAT36383.1 ADMH02000048 ETN67992.1 KZ288292 PBC29091.1 JXUM01030890 KQ560901 KXJ80422.1 GBYB01001520 GBYB01001521 GBYB01001522 GBYB01001523 GBYB01007951 GBYB01011553 JAG71287.1 JAG71288.1 JAG71289.1 JAG71290.1 JAG77718.1 JAG81320.1 AXCM01001491 ATLV01016361 KE525079 KFB41310.1 AAZO01005147 DS235751 EEB16384.1 GEZM01046474 JAV77210.1 GL732542 EFX81697.1 QOIP01000007 RLU20378.1 KK107364 EZA51939.1 APCN01002661 KQ977408 KYN02960.1 GGFK01010806 MBW44127.1 ADTU01011962 ADTU01011963 ADTU01011964 NNAY01001737 OXU23077.1 CVRI01000021 CRK91496.1 GGFL01004600 MBW68778.1 GDUN01000236 JAN95683.1 GGFK01003762 MBW37083.1 GGFK01003689 MBW37010.1 GDIP01090024 JAM13691.1 GDIP01125922 JAL77792.1 GDIP01231964 JAI91437.1 GDIQ01033471 JAN61266.1 GDIQ01169244 JAK82481.1 GDIQ01150751 JAL00975.1 KQ982490 KYQ55807.1 KQ981727 KYN36952.1 CH933806 EDW15641.2 GDIP01239663 JAI83738.1 LJIG01001402 KRT85538.1 KQ976396 KYM93005.1 GDIQ01165505 JAK86220.1 GDIP01207218 JAJ16184.1 KQ978691 KYN29222.1 GDIP01022404 JAM81311.1 GDIP01215707 JAJ07695.1 GDIP01114019 JAL89695.1 LRGB01001348 KZS12822.1 GDIQ01113114 JAL38612.1 GDIQ01132462 JAL19264.1 GDIQ01132463 JAL19263.1 GDIQ01007414 JAN87323.1 GDIQ01073034 JAN21703.1 GDIP01038685 JAM65030.1 CH940650 EDW66854.2 AAAB01008978 EGK97364.1 CH916369 EDV92549.1 CH964272 EDW85097.1 GALX01001278 JAB67188.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000053240
UP000235965
+ More
UP000053105 UP000053825 UP000027135 UP000000311 UP000092462 UP000076502 UP000192223 UP000092461 UP000036403 UP000007266 UP000008237 UP000007755 UP000030742 UP000019118 UP000002320 UP000245037 UP000075880 UP000008820 UP000000673 UP000242457 UP000005203 UP000069940 UP000249989 UP000075883 UP000069272 UP000075900 UP000030765 UP000009046 UP000075885 UP000076408 UP000000305 UP000279307 UP000053097 UP000076407 UP000075902 UP000075920 UP000075882 UP000075840 UP000078542 UP000075903 UP000075884 UP000075881 UP000002358 UP000005205 UP000215335 UP000183832 UP000075809 UP000078541 UP000009192 UP000078540 UP000078492 UP000076858 UP000008792 UP000007062 UP000001070 UP000007798 UP000192221
UP000053105 UP000053825 UP000027135 UP000000311 UP000092462 UP000076502 UP000192223 UP000092461 UP000036403 UP000007266 UP000008237 UP000007755 UP000030742 UP000019118 UP000002320 UP000245037 UP000075880 UP000008820 UP000000673 UP000242457 UP000005203 UP000069940 UP000249989 UP000075883 UP000069272 UP000075900 UP000030765 UP000009046 UP000075885 UP000076408 UP000000305 UP000279307 UP000053097 UP000076407 UP000075902 UP000075920 UP000075882 UP000075840 UP000078542 UP000075903 UP000075884 UP000075881 UP000002358 UP000005205 UP000215335 UP000183832 UP000075809 UP000078541 UP000009192 UP000078540 UP000078492 UP000076858 UP000008792 UP000007062 UP000001070 UP000007798 UP000192221
PRIDE
Interpro
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
H9J621
A0A2H1VG59
A0A2A4K449
A0A194PGA1
A0A212FIM0
A0A194RLH2
+ More
A0A2W1BIW6 A0A2J7PR16 A0A1B6MBW9 A0A1B6DEG6 A0A0M9A4E5 A0A0L7QME1 A0A067R9P9 E2AK71 A0A1B0D9S2 A0A1B6HYD6 A0A154PIH9 A0A1W4WW62 A0A1B0CCV8 A0A1B6ED22 A0A0J7NUM9 A0A1B0GNZ8 E9IMC0 A0A139WG03 E2BAV8 A0A1B6EMU7 A0A1B6GWX1 F4WFF0 A0A0C9QDX1 U4U6K8 N6SSY4 B0W921 A0A2P8ZK85 A0A182JI35 Q16PU5 W5JUG5 A0A2A3EDJ5 A0A087ZT71 A0A182HA09 A0A0C9QLK7 A0A182MNY7 A0A182FA40 A0A182RFK2 A0A084VTL6 E0VSM8 A0A182P426 A0A182YG14 A0A1Y1LXY0 E9GFT7 A0A3L8DKL5 A0A026WA84 A0A182XTK9 A0A182TXC2 A0A182VWZ2 A0A182KMM2 A0A182I873 A0A195CQN9 A0A2M4ATM6 A0A182USE3 A0A182NDY5 A0A182JSW3 K7IQG0 A0A158NCM0 A0A232EXP4 A0A1J1HTU8 A0A2M4CVB5 A0A0P6J137 A0A2M4A8F1 A0A2M4A8E8 A0A0P5W7X3 A0A0P5TQM9 A0A0P4YCV3 A0A0P6HES3 A0A0P5LI99 A0A0P5MV71 A0A151X614 A0A195F9R1 B4K9Z3 A0A0P4XLG9 A0A0T6BDX9 A0A195BXI1 A0A0P5LU86 A0A0P5A7Y0 A0A195EMU4 A0A0P6B471 A0A0P4Z9N0 A0A0P5UI14 A0A162C8N0 A0A0P5R972 A0A0P5P7U8 A0A0P5P7T0 A0A0P6JI64 A0A0P6DTT8 A0A0P5ZUZ1 B4LXY8 F5HM53 B4JG10 B4NK55 V5H2X1 A0A1W4WBZ9
A0A2W1BIW6 A0A2J7PR16 A0A1B6MBW9 A0A1B6DEG6 A0A0M9A4E5 A0A0L7QME1 A0A067R9P9 E2AK71 A0A1B0D9S2 A0A1B6HYD6 A0A154PIH9 A0A1W4WW62 A0A1B0CCV8 A0A1B6ED22 A0A0J7NUM9 A0A1B0GNZ8 E9IMC0 A0A139WG03 E2BAV8 A0A1B6EMU7 A0A1B6GWX1 F4WFF0 A0A0C9QDX1 U4U6K8 N6SSY4 B0W921 A0A2P8ZK85 A0A182JI35 Q16PU5 W5JUG5 A0A2A3EDJ5 A0A087ZT71 A0A182HA09 A0A0C9QLK7 A0A182MNY7 A0A182FA40 A0A182RFK2 A0A084VTL6 E0VSM8 A0A182P426 A0A182YG14 A0A1Y1LXY0 E9GFT7 A0A3L8DKL5 A0A026WA84 A0A182XTK9 A0A182TXC2 A0A182VWZ2 A0A182KMM2 A0A182I873 A0A195CQN9 A0A2M4ATM6 A0A182USE3 A0A182NDY5 A0A182JSW3 K7IQG0 A0A158NCM0 A0A232EXP4 A0A1J1HTU8 A0A2M4CVB5 A0A0P6J137 A0A2M4A8F1 A0A2M4A8E8 A0A0P5W7X3 A0A0P5TQM9 A0A0P4YCV3 A0A0P6HES3 A0A0P5LI99 A0A0P5MV71 A0A151X614 A0A195F9R1 B4K9Z3 A0A0P4XLG9 A0A0T6BDX9 A0A195BXI1 A0A0P5LU86 A0A0P5A7Y0 A0A195EMU4 A0A0P6B471 A0A0P4Z9N0 A0A0P5UI14 A0A162C8N0 A0A0P5R972 A0A0P5P7U8 A0A0P5P7T0 A0A0P6JI64 A0A0P6DTT8 A0A0P5ZUZ1 B4LXY8 F5HM53 B4JG10 B4NK55 V5H2X1 A0A1W4WBZ9
PDB
6QDV
E-value=1.06425e-86,
Score=814
Ontologies
GO
PANTHER
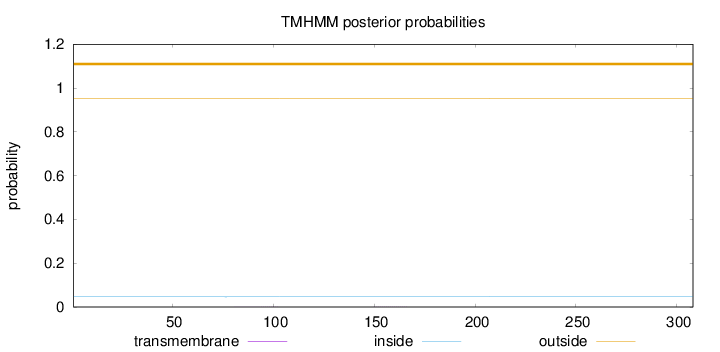
Topology
Subcellular location
Nucleus
Length:
308
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00745000000000001
Exp number, first 60 AAs:
0.00017
Total prob of N-in:
0.04650
outside
1 - 308
Population Genetic Test Statistics
Pi
338.906386
Theta
172.32977
Tajima's D
2.043552
CLR
0
CSRT
0.887555622218889
Interpretation
Uncertain
