Gene
KWMTBOMO15208
Pre Gene Modal
BGIBMGA004952
Annotation
PREDICTED:_probable_G-protein_coupled_receptor_Mth-like_3_[Papilio_xuthus]
Location in the cell
PlasmaMembrane Reliability : 4.834
Sequence
CDS
ATGAACGTGATTTGTATAAACGTATGTCGTTCAGTGCTCAACGCTTCTCGCTTGGCAAAATCGGATAGAAAGCAATTTCTGTGGTACAGTTGTTACGCGTGGGGCGGTACTTTAGCACTACTCTTTGTGGCCATCATTACGACCTTCGTCGACGGAGACCACTGGAAACCAGGTTTTGGTGAATCGAGTTGTTGGTTCAACGGTCGCACCGAAACATGGATCTTTTTCTACGGACCGATTGCGTGTTTGATAGCGTTGAATATTGGCCTATTCTTGTTGTCATTTGTTCATTTGTGGCTGCAGACTATTAAATACGAAGTCAATAAACTGAACAACTTGAAGTATAAATTCCTGCTATCATTGAAACTCTTTTTGGTCATGGGGATATCCTGGATATTTGAAATCGCCAGTTTCGCTCACGGAAAGTCGCATATTATCTGGAAAATCATGGATATTTTCAACTGCCTCCAAGGCGTTATTATATTCTTATTGCTCGTAGTGCTGAGAAAGAAAGTAATCCAAAATCTAGCGAATGAAAACTGTTGTGTTTTCTTAACACGCCGGCTCTCGGATAAACTTAGCCCACATGATGATTCCGATGATCAACAGAACATACTGGGCGATGAAACCGTCGAAGTCCGATTAAATTAA
Protein
MNVICINVCRSVLNASRLAKSDRKQFLWYSCYAWGGTLALLFVAIITTFVDGDHWKPGFGESSCWFNGRTETWIFFYGPIACLIALNIGLFLLSFVHLWLQTIKYEVNKLNNLKYKFLLSLKLFLVMGISWIFEIASFAHGKSHIIWKIMDIFNCLQGVIIFLLLVVLRKKVIQNLANENCCVFLTRRLSDKLSPHDDSDDQQNILGDETVEVRLN
Summary
Uniprot
A0A194RK10
A0A2A4K4G0
A0A194PMI3
S4PCU8
A0A212FIL6
A0A2H1VCN5
+ More
A0A2W1BJF1 A0A0N0BI45 A0A2A3EBE7 V9ICZ7 A0A087ZTD3 A0A154PHJ8 A0A0L7QM33 F4WFE8 A0A195EMW3 A0A195CR44 E2BAW0 A0A195BWP8 A0A158NCL8 A0A151X679 A0A195F936 A0A026W8G7 A0A0J7KW15 A0A232EXJ5 A0A1E1W8Q0 K7JNF9 E2AK69 A0A1Y1LVI2 A0A1B6CZE8 A0A1B6CQ85 A0A1B6DH68 A0A1B6MAP6 A0A0R7EYQ8 A0A2J7PR29 A0A2A4JL21 A0A1W4WKC5 A0A0P4VSE1 A0A1B6HCF7 A0A1B6J325 A0A1B6G4C0 A0A224XDM6 A0A0P4VVT9 A0A1B0CCW0 E0VYH0 A0A0T6B6U3 A0A194PH00 V5I9E6 A0A139WG45 A0A1L8DJZ7 A0A1L8DJS0 A0A1B0D8R9 A0A2J7QUS9 A0A194RL71 A0A212EHI9 A0A1Y1LEX7 A0A0A7P873 A0A2J7R7H5 A0A212F740 A0A2R7W2G3 A0A2P8ZK86 A0A1B6F3W1 A0A1B6F351 A0A224XRP7 A0A1B6JEQ5 A0A1W4X4A5 A0A1B6JBW8 A0A2R7WWB7 A0A1B6IBM7 A0A1B6IUT9 A0A1B6LT28 A0A0C9R8N0 A0A1B6KQY4 A0A0A1WH47 A0A0A1WL34 A0A0A1XR13 A0A067QGP0 D6X2H9 A0A1B6M8D2 A0A3L8DAZ6 A0A1B6G5F1 A0A336KT42 A0A336LR90 B3MQ68 A0A1W4VJ42 A0A2J7R7M9 J9K693 A0A2H8TZ60 A0A067RCC7 A0A2S2NLW8 A0A034WMS8 A0A0A1X956 A0A0P5E0J6 B4IEQ5 A0A1B0G4S0 A0A0P5K667 A0A0P5IYH2
A0A2W1BJF1 A0A0N0BI45 A0A2A3EBE7 V9ICZ7 A0A087ZTD3 A0A154PHJ8 A0A0L7QM33 F4WFE8 A0A195EMW3 A0A195CR44 E2BAW0 A0A195BWP8 A0A158NCL8 A0A151X679 A0A195F936 A0A026W8G7 A0A0J7KW15 A0A232EXJ5 A0A1E1W8Q0 K7JNF9 E2AK69 A0A1Y1LVI2 A0A1B6CZE8 A0A1B6CQ85 A0A1B6DH68 A0A1B6MAP6 A0A0R7EYQ8 A0A2J7PR29 A0A2A4JL21 A0A1W4WKC5 A0A0P4VSE1 A0A1B6HCF7 A0A1B6J325 A0A1B6G4C0 A0A224XDM6 A0A0P4VVT9 A0A1B0CCW0 E0VYH0 A0A0T6B6U3 A0A194PH00 V5I9E6 A0A139WG45 A0A1L8DJZ7 A0A1L8DJS0 A0A1B0D8R9 A0A2J7QUS9 A0A194RL71 A0A212EHI9 A0A1Y1LEX7 A0A0A7P873 A0A2J7R7H5 A0A212F740 A0A2R7W2G3 A0A2P8ZK86 A0A1B6F3W1 A0A1B6F351 A0A224XRP7 A0A1B6JEQ5 A0A1W4X4A5 A0A1B6JBW8 A0A2R7WWB7 A0A1B6IBM7 A0A1B6IUT9 A0A1B6LT28 A0A0C9R8N0 A0A1B6KQY4 A0A0A1WH47 A0A0A1WL34 A0A0A1XR13 A0A067QGP0 D6X2H9 A0A1B6M8D2 A0A3L8DAZ6 A0A1B6G5F1 A0A336KT42 A0A336LR90 B3MQ68 A0A1W4VJ42 A0A2J7R7M9 J9K693 A0A2H8TZ60 A0A067RCC7 A0A2S2NLW8 A0A034WMS8 A0A0A1X956 A0A0P5E0J6 B4IEQ5 A0A1B0G4S0 A0A0P5K667 A0A0P5IYH2
Pubmed
EMBL
KQ460045
KPJ18168.1
NWSH01000170
PCG78798.1
KQ459604
KPI92325.1
+ More
GAIX01003961 JAA88599.1 AGBW02008360 OWR53576.1 ODYU01001835 SOQ38575.1 KZ150014 PZC74998.1 KQ435736 KOX77022.1 KZ288292 PBC29093.1 JR038599 JR038600 AEY58336.1 AEY58337.1 KQ434905 KZC11292.1 KQ414894 KOC59703.1 GL888115 EGI67199.1 KQ978691 KYN29224.1 KQ977408 KYN02962.1 GL446861 EFN87167.1 KQ976396 KYM93007.1 ADTU01011962 ADTU01011963 KQ982490 KYQ55809.1 KQ981727 KYN36953.1 KK107364 QOIP01000007 EZA51941.1 RLU20375.1 LBMM01002673 KMQ94493.1 NNAY01001737 OXU23081.1 GDQN01007710 GDQN01001310 GDQN01001080 GDQN01000262 JAT83344.1 JAT89744.1 JAT89974.1 JAT90792.1 GL440194 EFN66173.1 GEZM01049214 JAV76035.1 GEDC01031549 GEDC01018478 JAS05749.1 JAS18820.1 GEDC01021738 JAS15560.1 GEDC01027779 GEDC01012306 JAS09519.1 JAS24992.1 GEBQ01006976 JAT33001.1 KP296186 AKN80718.1 NEVH01022633 PNF18784.1 NWSH01001119 PCG72509.1 GDKW01002665 JAI53930.1 GECU01035346 JAS72360.1 GECU01014117 JAS93589.1 GECZ01012492 JAS57277.1 GFTR01005889 JAW10537.1 GDRN01092840 JAI60025.1 AJWK01007142 AJWK01007143 DS235845 EEB18426.1 LJIG01009442 KRT83071.1 KPI92323.1 GALX01003070 JAB65396.1 KQ971351 KYB26825.1 GFDF01007387 JAV06697.1 GFDF01007388 JAV06696.1 AJVK01027681 AJVK01027682 NEVH01010486 PNF32336.1 KPJ18169.1 AGBW02014878 OWR40949.1 GEZM01062645 JAV69567.1 KM588897 AJA06111.1 NEVH01006732 PNF36788.1 AGBW02009936 OWR49550.1 KK854242 PTY13618.1 PYGN01000032 PSN56908.1 GECZ01024998 JAS44771.1 GECZ01025080 JAS44689.1 GFTR01003928 JAW12498.1 GECU01010087 JAS97619.1 GECU01011109 GECU01002381 JAS96597.1 JAT05326.1 KK855769 PTY23838.1 GECU01023436 JAS84270.1 GECU01017005 JAS90701.1 GEBQ01013134 JAT26843.1 GBYB01012724 JAG82491.1 GEBQ01026124 JAT13853.1 GBXI01016271 JAC98020.1 GBXI01014715 JAC99576.1 GBXI01000826 JAD13466.1 KK853497 KDR07201.1 KQ971372 EFA10672.1 GEBQ01007873 JAT32104.1 QOIP01000010 RLU17494.1 GECZ01012105 JAS57664.1 UFQS01000912 UFQT01000912 SSX07619.1 SSX27957.1 UFQS01000047 UFQT01000047 SSW98531.1 SSX18917.1 CH902621 EDV44494.1 NEVH01006727 PNF36848.1 ABLF02039939 GFXV01007722 MBW19527.1 KK852556 KDR21417.1 GGMR01005157 MBY17776.1 GAKP01003874 JAC55078.1 GBXI01007084 JAD07208.1 GDIP01149638 JAJ73764.1 CH480832 EDW46159.1 CCAG010019908 GDIQ01251880 GDIQ01250436 GDIQ01208952 GDIQ01207653 GDIQ01206246 GDIQ01190154 JAK61571.1 GDIQ01251881 GDIQ01250437 GDIQ01207654 GDIQ01206244 GDIQ01188398 JAK44071.1
GAIX01003961 JAA88599.1 AGBW02008360 OWR53576.1 ODYU01001835 SOQ38575.1 KZ150014 PZC74998.1 KQ435736 KOX77022.1 KZ288292 PBC29093.1 JR038599 JR038600 AEY58336.1 AEY58337.1 KQ434905 KZC11292.1 KQ414894 KOC59703.1 GL888115 EGI67199.1 KQ978691 KYN29224.1 KQ977408 KYN02962.1 GL446861 EFN87167.1 KQ976396 KYM93007.1 ADTU01011962 ADTU01011963 KQ982490 KYQ55809.1 KQ981727 KYN36953.1 KK107364 QOIP01000007 EZA51941.1 RLU20375.1 LBMM01002673 KMQ94493.1 NNAY01001737 OXU23081.1 GDQN01007710 GDQN01001310 GDQN01001080 GDQN01000262 JAT83344.1 JAT89744.1 JAT89974.1 JAT90792.1 GL440194 EFN66173.1 GEZM01049214 JAV76035.1 GEDC01031549 GEDC01018478 JAS05749.1 JAS18820.1 GEDC01021738 JAS15560.1 GEDC01027779 GEDC01012306 JAS09519.1 JAS24992.1 GEBQ01006976 JAT33001.1 KP296186 AKN80718.1 NEVH01022633 PNF18784.1 NWSH01001119 PCG72509.1 GDKW01002665 JAI53930.1 GECU01035346 JAS72360.1 GECU01014117 JAS93589.1 GECZ01012492 JAS57277.1 GFTR01005889 JAW10537.1 GDRN01092840 JAI60025.1 AJWK01007142 AJWK01007143 DS235845 EEB18426.1 LJIG01009442 KRT83071.1 KPI92323.1 GALX01003070 JAB65396.1 KQ971351 KYB26825.1 GFDF01007387 JAV06697.1 GFDF01007388 JAV06696.1 AJVK01027681 AJVK01027682 NEVH01010486 PNF32336.1 KPJ18169.1 AGBW02014878 OWR40949.1 GEZM01062645 JAV69567.1 KM588897 AJA06111.1 NEVH01006732 PNF36788.1 AGBW02009936 OWR49550.1 KK854242 PTY13618.1 PYGN01000032 PSN56908.1 GECZ01024998 JAS44771.1 GECZ01025080 JAS44689.1 GFTR01003928 JAW12498.1 GECU01010087 JAS97619.1 GECU01011109 GECU01002381 JAS96597.1 JAT05326.1 KK855769 PTY23838.1 GECU01023436 JAS84270.1 GECU01017005 JAS90701.1 GEBQ01013134 JAT26843.1 GBYB01012724 JAG82491.1 GEBQ01026124 JAT13853.1 GBXI01016271 JAC98020.1 GBXI01014715 JAC99576.1 GBXI01000826 JAD13466.1 KK853497 KDR07201.1 KQ971372 EFA10672.1 GEBQ01007873 JAT32104.1 QOIP01000010 RLU17494.1 GECZ01012105 JAS57664.1 UFQS01000912 UFQT01000912 SSX07619.1 SSX27957.1 UFQS01000047 UFQT01000047 SSW98531.1 SSX18917.1 CH902621 EDV44494.1 NEVH01006727 PNF36848.1 ABLF02039939 GFXV01007722 MBW19527.1 KK852556 KDR21417.1 GGMR01005157 MBY17776.1 GAKP01003874 JAC55078.1 GBXI01007084 JAD07208.1 GDIP01149638 JAJ73764.1 CH480832 EDW46159.1 CCAG010019908 GDIQ01251880 GDIQ01250436 GDIQ01208952 GDIQ01207653 GDIQ01206246 GDIQ01190154 JAK61571.1 GDIQ01251881 GDIQ01250437 GDIQ01207654 GDIQ01206244 GDIQ01188398 JAK44071.1
Proteomes
UP000053240
UP000218220
UP000053268
UP000007151
UP000053105
UP000242457
+ More
UP000005203 UP000076502 UP000053825 UP000007755 UP000078492 UP000078542 UP000008237 UP000078540 UP000005205 UP000075809 UP000078541 UP000053097 UP000279307 UP000036403 UP000215335 UP000002358 UP000000311 UP000203433 UP000235965 UP000192223 UP000092461 UP000009046 UP000007266 UP000092462 UP000245037 UP000027135 UP000007801 UP000192221 UP000007819 UP000001292 UP000092444
UP000005203 UP000076502 UP000053825 UP000007755 UP000078492 UP000078542 UP000008237 UP000078540 UP000005205 UP000075809 UP000078541 UP000053097 UP000279307 UP000036403 UP000215335 UP000002358 UP000000311 UP000203433 UP000235965 UP000192223 UP000092461 UP000009046 UP000007266 UP000092462 UP000245037 UP000027135 UP000007801 UP000192221 UP000007819 UP000001292 UP000092444
Interpro
SUPFAM
SSF63877
SSF63877
Gene 3D
ProteinModelPortal
A0A194RK10
A0A2A4K4G0
A0A194PMI3
S4PCU8
A0A212FIL6
A0A2H1VCN5
+ More
A0A2W1BJF1 A0A0N0BI45 A0A2A3EBE7 V9ICZ7 A0A087ZTD3 A0A154PHJ8 A0A0L7QM33 F4WFE8 A0A195EMW3 A0A195CR44 E2BAW0 A0A195BWP8 A0A158NCL8 A0A151X679 A0A195F936 A0A026W8G7 A0A0J7KW15 A0A232EXJ5 A0A1E1W8Q0 K7JNF9 E2AK69 A0A1Y1LVI2 A0A1B6CZE8 A0A1B6CQ85 A0A1B6DH68 A0A1B6MAP6 A0A0R7EYQ8 A0A2J7PR29 A0A2A4JL21 A0A1W4WKC5 A0A0P4VSE1 A0A1B6HCF7 A0A1B6J325 A0A1B6G4C0 A0A224XDM6 A0A0P4VVT9 A0A1B0CCW0 E0VYH0 A0A0T6B6U3 A0A194PH00 V5I9E6 A0A139WG45 A0A1L8DJZ7 A0A1L8DJS0 A0A1B0D8R9 A0A2J7QUS9 A0A194RL71 A0A212EHI9 A0A1Y1LEX7 A0A0A7P873 A0A2J7R7H5 A0A212F740 A0A2R7W2G3 A0A2P8ZK86 A0A1B6F3W1 A0A1B6F351 A0A224XRP7 A0A1B6JEQ5 A0A1W4X4A5 A0A1B6JBW8 A0A2R7WWB7 A0A1B6IBM7 A0A1B6IUT9 A0A1B6LT28 A0A0C9R8N0 A0A1B6KQY4 A0A0A1WH47 A0A0A1WL34 A0A0A1XR13 A0A067QGP0 D6X2H9 A0A1B6M8D2 A0A3L8DAZ6 A0A1B6G5F1 A0A336KT42 A0A336LR90 B3MQ68 A0A1W4VJ42 A0A2J7R7M9 J9K693 A0A2H8TZ60 A0A067RCC7 A0A2S2NLW8 A0A034WMS8 A0A0A1X956 A0A0P5E0J6 B4IEQ5 A0A1B0G4S0 A0A0P5K667 A0A0P5IYH2
A0A2W1BJF1 A0A0N0BI45 A0A2A3EBE7 V9ICZ7 A0A087ZTD3 A0A154PHJ8 A0A0L7QM33 F4WFE8 A0A195EMW3 A0A195CR44 E2BAW0 A0A195BWP8 A0A158NCL8 A0A151X679 A0A195F936 A0A026W8G7 A0A0J7KW15 A0A232EXJ5 A0A1E1W8Q0 K7JNF9 E2AK69 A0A1Y1LVI2 A0A1B6CZE8 A0A1B6CQ85 A0A1B6DH68 A0A1B6MAP6 A0A0R7EYQ8 A0A2J7PR29 A0A2A4JL21 A0A1W4WKC5 A0A0P4VSE1 A0A1B6HCF7 A0A1B6J325 A0A1B6G4C0 A0A224XDM6 A0A0P4VVT9 A0A1B0CCW0 E0VYH0 A0A0T6B6U3 A0A194PH00 V5I9E6 A0A139WG45 A0A1L8DJZ7 A0A1L8DJS0 A0A1B0D8R9 A0A2J7QUS9 A0A194RL71 A0A212EHI9 A0A1Y1LEX7 A0A0A7P873 A0A2J7R7H5 A0A212F740 A0A2R7W2G3 A0A2P8ZK86 A0A1B6F3W1 A0A1B6F351 A0A224XRP7 A0A1B6JEQ5 A0A1W4X4A5 A0A1B6JBW8 A0A2R7WWB7 A0A1B6IBM7 A0A1B6IUT9 A0A1B6LT28 A0A0C9R8N0 A0A1B6KQY4 A0A0A1WH47 A0A0A1WL34 A0A0A1XR13 A0A067QGP0 D6X2H9 A0A1B6M8D2 A0A3L8DAZ6 A0A1B6G5F1 A0A336KT42 A0A336LR90 B3MQ68 A0A1W4VJ42 A0A2J7R7M9 J9K693 A0A2H8TZ60 A0A067RCC7 A0A2S2NLW8 A0A034WMS8 A0A0A1X956 A0A0P5E0J6 B4IEQ5 A0A1B0G4S0 A0A0P5K667 A0A0P5IYH2
Ontologies
GO
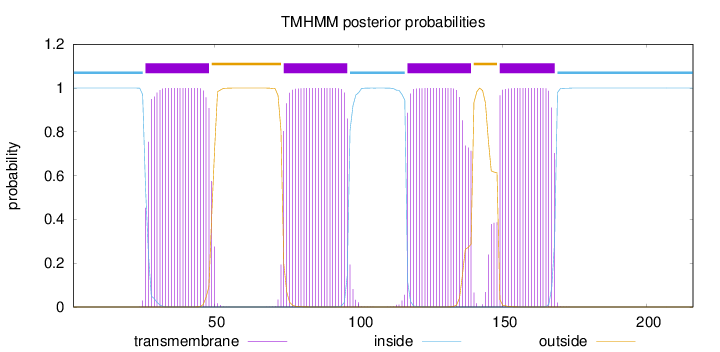
Topology
Length:
216
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
88.99533
Exp number, first 60 AAs:
22.8481
Total prob of N-in:
1.00000
POSSIBLE N-term signal
sequence
inside
1 - 25
TMhelix
26 - 48
outside
49 - 73
TMhelix
74 - 96
inside
97 - 116
TMhelix
117 - 139
outside
140 - 148
TMhelix
149 - 168
inside
169 - 216
Population Genetic Test Statistics
Pi
231.235998
Theta
199.055134
Tajima's D
1.030855
CLR
0.116679
CSRT
0.663766811659417
Interpretation
Uncertain
