Pre Gene Modal
BGIBMGA004963
Annotation
PREDICTED:_intron-binding_protein_aquarius_isoform_X2_[Bombyx_mori]
Full name
RNA helicase aquarius
Location in the cell
Cytoplasmic Reliability : 1.469 Mitochondrial Reliability : 1.194 Nuclear Reliability : 1.61
Sequence
CDS
ATGCTGAAGTTTTACGCAAGATTTGAGATTAGCGATCAGACCGGAGATCCTATGACCGACCGTGACATGACATTACAGCACTACTCAAAAATCACCTCACTACAGAAAGCCGCATTTTCCAAGTTTCCTGATCTTCGTTTATTTGCCCTCGCGAATGTAGCGAGTGTCGACACCAGAGAAACGTTGCAGAAGCACTTCAAGAATTTGAGTGACAAAGCTTTAAGAGCAATCGCCACGTACCTGAACCTGGTTCCTCCTGAAGGAAAAGAAGACGAGACGCCTTGGCACCGACTCGACAAAGATTTCCTCAGAGAGCTATTGATCTCGCGTCACGAGAGGCGAATTTCACAATTGGAAGAATTGAACTTAATGCCCCTGTATCCTACAGAAGAGATTATCTGGGACGAGAATGTAGTGCCCACAGAGATATACAGCGGAGAGAATTGCCTAGCCCTGCCTAAGCTGAATCTACAATTTTTAACATTACACGACTATCTGTTACGAAACTTCAATTTGTTCCGCTTGGAGAGCACATACGAGATAAGACAAGACATTGAAGACGCCGTATACAGATTGGCGCCGTGGAATTCAGAAGACGGAAGCGTTTATTTTGGGGGCTGGGCGCGGATGGCGCATCCTATAAAGAGCTTCGCAGTTGTCGAAGTGGCCAAACCGAACATCGGCGAGAAGGCCCCGTCGAGGGTCCGAGCCGATGTCACCGTCACGTTGAGCCTCAGAAACGAGATAAAACACGAATGGGAAAACCTCAGGAAGCATGACGTTTGCTTCCTGGTAACCGTCAGACCTACACAAGGCATAGGTACAAAGTATGACTACAGAAAGAGTATGGTGGATCAGGCGGGGATAGTGTACGTGAGGGGTTGCGAAGTTGAAGGTATGCTGGACGCATCCGGCAGAGTCATTGAGGACGGGCCGGAACCGCGGCCGGAGTTAGAGGGAGATTCCAGGACCTTCAGGCTCTTACTCGATCCTAACCAATACAGACTTGACTTGGACAGAGCTAGTAAAGGGAATGAGGACGTGTACGAGACTTTCAATATAGTTATGCGTCGTAAACCGAAAGAGAACAACTTCAAGGCCGTATTGGAAACGATACGAGAATTAATGAACACCGAATGCGTTGTACCGGACTGGCTTCACGATATAGTCTTGGGCTACGGAGATCCCGGTCAAGCTCATTACACGAGGATGCCGAACGAGATAGCTACTCTGGACTTCAACGATACATTCCTGAACATGGATCATCTGCGGAACAGCTTCCCGGGGTACGAAATCAAGGTGCAGACCAACGACCCAAGGAAGCTCGTCAGGCCGTTCAAGTTGACTTTCGAAAACGTCGTACGGAAGCAGCAGGTCGGGGACGCAGCTATGGACGAAGACGAACCCCGCAAAGCGATCATTGTCGAACCCCACGTACAACCGAAGAGGGGACCGTATTTGTACAACGAACCAAAGAAGAATAATATACTATTCACTCCGACACAAGTGGAAGCTATACGTTCTGGAATGCAACCTGGACTCACATTGGTCGTCGGTCCACCTGGAACTGGTAAAACGGACGTGGCAGTTCAAATAATTTCGAACATATACCACAACTTCCCGTGGCAACGTACCCTGGTCGTAACGCACAGCAACCAGGCCCTGAATCAGCTATTCGAGAAGGTCGCCGAGCTGGATGTAGACGAAAGGCATCTACTGCGTCTCGGTCACGGCGAGGAAGCTCTTCAGACAGACAAAGACTTCTCCAGATACGGCCGCGTGAACTATGTGTTGGCAAAGCGTCTGGAGCTGCTCGAGCTGGTGTCGCGGCTGCAGCGAACGTTGGACGCGGGCTCGGACGCCGGCGCTACGTGCGAGCTGGCGCACCACTTCCACGTCTACCACGTGCGGCCGCGCTGGCGAGCCTTCCTCGAGAACTGCCAGCAACAGACAAACTCGATTGAGATAGTCAGCAAGGAGTTTCCGTTCCACGAATTCTTCGATGATGCCCCGAAACCCTTGTTCCTGAGGAAGTCTCACGAAGAGGACATGGAAATAGCCAAAAGCTGTTACAGGTATATTGATCACATATTCGAGGAATTGGAAGAGTTCCGAGCCTTCGAACTACTGAGGTCCGGACTGGATCGGTCCAAGTACCTGTTGGTCAAAGAAGCGAAAATAATCGCGATGACGTGCACGCACGCCGCCCTCAAACGTTCCGAATTAGTGCAAATGGGTTTCAAATACGACAACATACTGATGGAGGAATCAGCTCAAATCCTCGAGATCGAGACGTTCATACCGCTGCTCCTGCAGAACCCGCAGGACGGCAGGTCCCGGCTGAAGCGTTGGATCATGATCGGCGACCACCACCAGCTGCCCCCGGTCGTCAAGAACATGGCGTTCCAGAAGTACTGCAACATGGAACAGAGCCTGTTCACCAGAATGGTCAGACTGGGTGTTCCGTACGTGGAACTGGACGCACAAGGTCGAGCTAGACCCAGCATCTGCAACCTGTACCGCTGGCGCTACCGGGCGCTGGGCGACCTGTGCCACGTGACGCAGCTCCCGGAGTACCGCGCCGCCAACGCGGGCCTGCGACACGACTTCCAGCTCGTCAACGTGGACGACTTCAACGGCGTCGGCGAGACCGAGCCCAGCCCTTACTTCTATCAGAATTTAGCGGAAGCAGAATACGTTGTAGCGGTTTTCATGTACATGCGCCTGATCGGATGGCCTGCAGAGCGCATCTCCATACTGACCACATATAACGGGCAGAAACATCTCATCAGGGACGTCATAGATAAGCGATGTGCCGATAATCCACTCATCGGTCGACCTCACAAGGTGACGACGGTAGACAAGTACCAGGGCCAGCAGAACGACATAGCTCTAGTGTCGCTGGTCCGTACGAAGGCGGTGGGTCACGTGCGCGACCTGCGGAGACTCATCGTCGCCGCCTCAAGAGCCCGTCTCGGCCTATACATATTCGCGAGGGCCAACCTCTTCAGGAACTGCTACGAGCTACAGCCTACATTCAATCAGCTCGTGGAGCGTCCACTGCAGCTTGAACTAGTCCCGGGCGAGCGGTACCCGGCGCAGCGCGCGGCGGGCGTGGCCGTGCCGCCGGAGCTGCTGCTGTCCGTCCGGGACATGCCGCACATGGCGCGCTACGTCTACGACATGTACCTGCACATTGTCAACGACGCACCCGACGCTCGACAGAGTGAATAG
Protein
MLKFYARFEISDQTGDPMTDRDMTLQHYSKITSLQKAAFSKFPDLRLFALANVASVDTRETLQKHFKNLSDKALRAIATYLNLVPPEGKEDETPWHRLDKDFLRELLISRHERRISQLEELNLMPLYPTEEIIWDENVVPTEIYSGENCLALPKLNLQFLTLHDYLLRNFNLFRLESTYEIRQDIEDAVYRLAPWNSEDGSVYFGGWARMAHPIKSFAVVEVAKPNIGEKAPSRVRADVTVTLSLRNEIKHEWENLRKHDVCFLVTVRPTQGIGTKYDYRKSMVDQAGIVYVRGCEVEGMLDASGRVIEDGPEPRPELEGDSRTFRLLLDPNQYRLDLDRASKGNEDVYETFNIVMRRKPKENNFKAVLETIRELMNTECVVPDWLHDIVLGYGDPGQAHYTRMPNEIATLDFNDTFLNMDHLRNSFPGYEIKVQTNDPRKLVRPFKLTFENVVRKQQVGDAAMDEDEPRKAIIVEPHVQPKRGPYLYNEPKKNNILFTPTQVEAIRSGMQPGLTLVVGPPGTGKTDVAVQIISNIYHNFPWQRTLVVTHSNQALNQLFEKVAELDVDERHLLRLGHGEEALQTDKDFSRYGRVNYVLAKRLELLELVSRLQRTLDAGSDAGATCELAHHFHVYHVRPRWRAFLENCQQQTNSIEIVSKEFPFHEFFDDAPKPLFLRKSHEEDMEIAKSCYRYIDHIFEELEEFRAFELLRSGLDRSKYLLVKEAKIIAMTCTHAALKRSELVQMGFKYDNILMEESAQILEIETFIPLLLQNPQDGRSRLKRWIMIGDHHQLPPVVKNMAFQKYCNMEQSLFTRMVRLGVPYVELDAQGRARPSICNLYRWRYRALGDLCHVTQLPEYRAANAGLRHDFQLVNVDDFNGVGETEPSPYFYQNLAEAEYVVAVFMYMRLIGWPAERISILTTYNGQKHLIRDVIDKRCADNPLIGRPHKVTTVDKYQGQQNDIALVSLVRTKAVGHVRDLRRLIVAASRARLGLYIFARANLFRNCYELQPTFNQLVERPLQLELVPGERYPAQRAAGVAVPPELLLSVRDMPHMARYVYDMYLHIVNDAPDARQSE
Summary
Description
Intron-binding spliceosomal protein required to link pre-mRNA splicing and snoRNP (small nucleolar ribonucleoprotein) biogenesis. Plays a key role in position-dependent assembly of intron-encoded box C/D small snoRNP, splicing being required for snoRNP assembly. May act by helping the folding of the snoRNA sequence. Binds to intron of pre-mRNAs in a sequence-independent manner, contacting the region between snoRNA and the branchpoint of introns (40 nucleotides upstream of the branchpoint) during the late stages of splicing.
Subunit
Identified in the spliceosome C complex. Component of the XAB2 complex, a multimeric protein complex composed of XAB2, PRPF19, AQR, ZNF830, ISY1, and PPIE.
Similarity
Belongs to the CWF11 family.
Uniprot
H9J622
A0A2A4JLC6
A0A194PHX3
A0A194RLH7
A0A2W1BNZ4
A0A2J7PR08
+ More
A0A067R9P9 A0A232EXP4 K7IQG0 A0A087ZT71 A0A2A3EDJ5 A0A0M9A6N3 E2BAW2 A0A0L7QMC6 A0A0C9QLK7 A0A158NCM0 A0A151X614 F4WFE7 A0A195F9R1 A0A195EMU4 A0A195BXI1 E2AK68 E0VSM8 E9IMC2 A0A0J7KW62 A0A1W4XP01 A0A026WA84 A0A3L8DKL5 A0A154PHF7 A0A139WFX9 A0A195CQN9 A0A1B6KBV0 A0A1S4G0D7 A0A1Y1LXY0 Q16IB0 A0A0P6J137 A0A182G802 A0A034VGH3 A0A182LS08 A0A0K8VLN3 A0A182P429 A0A0A1WJY7 A0A182QEI5 A0A182VWZ5 A0A2M4A8F1 A0A2M4A9Q0 A0A182YG12 A0A2M4A8E8 A0A0L0CC12 B0W925 A0A182RFJ9 A0A182FA37 A0A182SJS1 A0A182NDY0 W5JKE2 A0A087Z6K8 A0A2M4CVB5 A0A182K5H0 A0A182U8A4 A0A182I870 W8AM97 F5HM53 A0A384D0C3 A0A3Q7UI82 A0A3Q2H0V9 L5K2K4 A0A286ZI68 A0A182XKI2 A0A2Y9HN94 A0A2K6S0F9 Q7Q257 A0A0N8ETY7 D2H7R7 W5QFI2 A0A2R9AK31 A0A096NTB7 A0A2K6RM46 M3WK08 F7H585 A0A091DR01 A0A096NTB5 A0A2U3XFX8 A0A2U3WJR2 A0A2K6S0G9 A0A3Q0D6E4 A0A2K5URQ7 L8IU75 A0A2K6AGR0 A0A0D9R489 A0A3Q7T2L7 A5D7E4 H9ZFS7 H3AVK7 A0A2U4BC65 A0A2R9AEP9 A0A2R9ADM6 A0A2K5NC12 A0A2J8JB32 A0A3Q7SDH2 A0A2K5KC27
A0A067R9P9 A0A232EXP4 K7IQG0 A0A087ZT71 A0A2A3EDJ5 A0A0M9A6N3 E2BAW2 A0A0L7QMC6 A0A0C9QLK7 A0A158NCM0 A0A151X614 F4WFE7 A0A195F9R1 A0A195EMU4 A0A195BXI1 E2AK68 E0VSM8 E9IMC2 A0A0J7KW62 A0A1W4XP01 A0A026WA84 A0A3L8DKL5 A0A154PHF7 A0A139WFX9 A0A195CQN9 A0A1B6KBV0 A0A1S4G0D7 A0A1Y1LXY0 Q16IB0 A0A0P6J137 A0A182G802 A0A034VGH3 A0A182LS08 A0A0K8VLN3 A0A182P429 A0A0A1WJY7 A0A182QEI5 A0A182VWZ5 A0A2M4A8F1 A0A2M4A9Q0 A0A182YG12 A0A2M4A8E8 A0A0L0CC12 B0W925 A0A182RFJ9 A0A182FA37 A0A182SJS1 A0A182NDY0 W5JKE2 A0A087Z6K8 A0A2M4CVB5 A0A182K5H0 A0A182U8A4 A0A182I870 W8AM97 F5HM53 A0A384D0C3 A0A3Q7UI82 A0A3Q2H0V9 L5K2K4 A0A286ZI68 A0A182XKI2 A0A2Y9HN94 A0A2K6S0F9 Q7Q257 A0A0N8ETY7 D2H7R7 W5QFI2 A0A2R9AK31 A0A096NTB7 A0A2K6RM46 M3WK08 F7H585 A0A091DR01 A0A096NTB5 A0A2U3XFX8 A0A2U3WJR2 A0A2K6S0G9 A0A3Q0D6E4 A0A2K5URQ7 L8IU75 A0A2K6AGR0 A0A0D9R489 A0A3Q7T2L7 A5D7E4 H9ZFS7 H3AVK7 A0A2U4BC65 A0A2R9AEP9 A0A2R9ADM6 A0A2K5NC12 A0A2J8JB32 A0A3Q7SDH2 A0A2K5KC27
Pubmed
19121390
26354079
28756777
24845553
28648823
20075255
+ More
20798317 21347285 21719571 20566863 21282665 24508170 30249741 18362917 19820115 17510324 28004739 26999592 26483478 25348373 25830018 25244985 26108605 20920257 24495485 12364791 14747013 17210077 24813606 19892987 23258410 20010809 20809919 22722832 25362486 17975172 22751099 25319552 9215903
20798317 21347285 21719571 20566863 21282665 24508170 30249741 18362917 19820115 17510324 28004739 26999592 26483478 25348373 25830018 25244985 26108605 20920257 24495485 12364791 14747013 17210077 24813606 19892987 23258410 20010809 20809919 22722832 25362486 17975172 22751099 25319552 9215903
EMBL
BABH01029882
BABH01029883
BABH01029884
BABH01029885
NWSH01001119
PCG72508.1
+ More
KQ459604 KPI92324.1 KQ460045 KPJ18170.1 KZ150014 PZC74997.1 NEVH01022633 PNF18773.1 KK852653 KDR19357.1 NNAY01001737 OXU23077.1 KZ288292 PBC29091.1 KQ435736 KOX77023.1 GL446861 EFN87169.1 KQ414894 KOC59704.1 GBYB01001520 GBYB01001521 GBYB01001522 GBYB01001523 GBYB01007951 GBYB01011553 JAG71287.1 JAG71288.1 JAG71289.1 JAG71290.1 JAG77718.1 JAG81320.1 ADTU01011962 ADTU01011963 ADTU01011964 KQ982490 KYQ55807.1 GL888115 EGI67198.1 KQ981727 KYN36952.1 KQ978691 KYN29222.1 KQ976396 KYM93005.1 GL440194 EFN66172.1 AAZO01005147 DS235751 EEB16384.1 GL764129 EFZ18539.1 LBMM01002673 KMQ94494.1 KK107364 EZA51939.1 QOIP01000007 RLU20378.1 KQ434905 KZC11291.1 KQ971351 KYB26824.1 KQ977408 KYN02960.1 GEBQ01031049 JAT08928.1 GEZM01046474 JAV77210.1 CH478091 EAT33999.1 GDUN01000236 JAN95683.1 JXUM01047201 KQ561510 KXJ78338.1 GAKP01018067 JAC40885.1 AXCM01002744 GDHF01012829 JAI39485.1 GBXI01015291 JAC99000.1 AXCN02002211 GGFK01003762 MBW37083.1 GGFK01004202 MBW37523.1 GGFK01003689 MBW37010.1 JRES01000630 KNC29790.1 DS231861 EDS39463.1 ADMH02001261 ETN63350.1 GGFL01004600 MBW68778.1 APCN01002659 GAMC01016635 JAB89920.1 AAAB01008978 EGK97364.1 KB031042 ELK05567.1 AEMK02000004 EAA13627.5 GEBF01003067 JAO00566.1 GL192565 EFB17996.1 AMGL01102452 AMGL01102453 AMGL01102454 AJFE02038412 AJFE02038413 AJFE02038414 AJFE02038415 AJFE02038416 AJFE02038417 AJFE02038418 AJFE02038419 AHZZ02030743 AHZZ02030744 AHZZ02030745 AANG04001761 KN122106 KFO32908.1 AQIA01062161 AQIA01062162 AQIA01062163 AQIA01062164 AQIA01062165 AQIA01062166 AQIA01062167 JH880809 ELR58707.1 AQIB01116411 AQIB01116412 AQIB01116413 BC140525 AAI40526.1 JU477989 AFH34793.1 AFYH01147097 AFYH01147098 AFYH01147099 AFYH01147100 AFYH01147101 AFYH01147102 AFYH01147103 AFYH01147104 AFYH01147105 NBAG03000488 PNI19963.1
KQ459604 KPI92324.1 KQ460045 KPJ18170.1 KZ150014 PZC74997.1 NEVH01022633 PNF18773.1 KK852653 KDR19357.1 NNAY01001737 OXU23077.1 KZ288292 PBC29091.1 KQ435736 KOX77023.1 GL446861 EFN87169.1 KQ414894 KOC59704.1 GBYB01001520 GBYB01001521 GBYB01001522 GBYB01001523 GBYB01007951 GBYB01011553 JAG71287.1 JAG71288.1 JAG71289.1 JAG71290.1 JAG77718.1 JAG81320.1 ADTU01011962 ADTU01011963 ADTU01011964 KQ982490 KYQ55807.1 GL888115 EGI67198.1 KQ981727 KYN36952.1 KQ978691 KYN29222.1 KQ976396 KYM93005.1 GL440194 EFN66172.1 AAZO01005147 DS235751 EEB16384.1 GL764129 EFZ18539.1 LBMM01002673 KMQ94494.1 KK107364 EZA51939.1 QOIP01000007 RLU20378.1 KQ434905 KZC11291.1 KQ971351 KYB26824.1 KQ977408 KYN02960.1 GEBQ01031049 JAT08928.1 GEZM01046474 JAV77210.1 CH478091 EAT33999.1 GDUN01000236 JAN95683.1 JXUM01047201 KQ561510 KXJ78338.1 GAKP01018067 JAC40885.1 AXCM01002744 GDHF01012829 JAI39485.1 GBXI01015291 JAC99000.1 AXCN02002211 GGFK01003762 MBW37083.1 GGFK01004202 MBW37523.1 GGFK01003689 MBW37010.1 JRES01000630 KNC29790.1 DS231861 EDS39463.1 ADMH02001261 ETN63350.1 GGFL01004600 MBW68778.1 APCN01002659 GAMC01016635 JAB89920.1 AAAB01008978 EGK97364.1 KB031042 ELK05567.1 AEMK02000004 EAA13627.5 GEBF01003067 JAO00566.1 GL192565 EFB17996.1 AMGL01102452 AMGL01102453 AMGL01102454 AJFE02038412 AJFE02038413 AJFE02038414 AJFE02038415 AJFE02038416 AJFE02038417 AJFE02038418 AJFE02038419 AHZZ02030743 AHZZ02030744 AHZZ02030745 AANG04001761 KN122106 KFO32908.1 AQIA01062161 AQIA01062162 AQIA01062163 AQIA01062164 AQIA01062165 AQIA01062166 AQIA01062167 JH880809 ELR58707.1 AQIB01116411 AQIB01116412 AQIB01116413 BC140525 AAI40526.1 JU477989 AFH34793.1 AFYH01147097 AFYH01147098 AFYH01147099 AFYH01147100 AFYH01147101 AFYH01147102 AFYH01147103 AFYH01147104 AFYH01147105 NBAG03000488 PNI19963.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000235965
UP000027135
+ More
UP000215335 UP000002358 UP000005203 UP000242457 UP000053105 UP000008237 UP000053825 UP000005205 UP000075809 UP000007755 UP000078541 UP000078492 UP000078540 UP000000311 UP000009046 UP000036403 UP000192223 UP000053097 UP000279307 UP000076502 UP000007266 UP000078542 UP000008820 UP000069940 UP000249989 UP000075883 UP000075885 UP000075886 UP000075920 UP000076408 UP000037069 UP000002320 UP000075900 UP000069272 UP000075901 UP000075884 UP000000673 UP000075881 UP000075902 UP000075840 UP000007062 UP000261680 UP000291021 UP000286642 UP000002281 UP000010552 UP000008227 UP000076407 UP000248481 UP000233220 UP000002356 UP000240080 UP000028761 UP000233200 UP000011712 UP000008225 UP000028990 UP000245341 UP000245340 UP000189706 UP000233100 UP000233140 UP000029965 UP000286640 UP000008672 UP000245320 UP000233060 UP000233080
UP000215335 UP000002358 UP000005203 UP000242457 UP000053105 UP000008237 UP000053825 UP000005205 UP000075809 UP000007755 UP000078541 UP000078492 UP000078540 UP000000311 UP000009046 UP000036403 UP000192223 UP000053097 UP000279307 UP000076502 UP000007266 UP000078542 UP000008820 UP000069940 UP000249989 UP000075883 UP000075885 UP000075886 UP000075920 UP000076408 UP000037069 UP000002320 UP000075900 UP000069272 UP000075901 UP000075884 UP000000673 UP000075881 UP000075902 UP000075840 UP000007062 UP000261680 UP000291021 UP000286642 UP000002281 UP000010552 UP000008227 UP000076407 UP000248481 UP000233220 UP000002356 UP000240080 UP000028761 UP000233200 UP000011712 UP000008225 UP000028990 UP000245341 UP000245340 UP000189706 UP000233100 UP000233140 UP000029965 UP000286640 UP000008672 UP000245320 UP000233060 UP000233080
PRIDE
Interpro
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
H9J622
A0A2A4JLC6
A0A194PHX3
A0A194RLH7
A0A2W1BNZ4
A0A2J7PR08
+ More
A0A067R9P9 A0A232EXP4 K7IQG0 A0A087ZT71 A0A2A3EDJ5 A0A0M9A6N3 E2BAW2 A0A0L7QMC6 A0A0C9QLK7 A0A158NCM0 A0A151X614 F4WFE7 A0A195F9R1 A0A195EMU4 A0A195BXI1 E2AK68 E0VSM8 E9IMC2 A0A0J7KW62 A0A1W4XP01 A0A026WA84 A0A3L8DKL5 A0A154PHF7 A0A139WFX9 A0A195CQN9 A0A1B6KBV0 A0A1S4G0D7 A0A1Y1LXY0 Q16IB0 A0A0P6J137 A0A182G802 A0A034VGH3 A0A182LS08 A0A0K8VLN3 A0A182P429 A0A0A1WJY7 A0A182QEI5 A0A182VWZ5 A0A2M4A8F1 A0A2M4A9Q0 A0A182YG12 A0A2M4A8E8 A0A0L0CC12 B0W925 A0A182RFJ9 A0A182FA37 A0A182SJS1 A0A182NDY0 W5JKE2 A0A087Z6K8 A0A2M4CVB5 A0A182K5H0 A0A182U8A4 A0A182I870 W8AM97 F5HM53 A0A384D0C3 A0A3Q7UI82 A0A3Q2H0V9 L5K2K4 A0A286ZI68 A0A182XKI2 A0A2Y9HN94 A0A2K6S0F9 Q7Q257 A0A0N8ETY7 D2H7R7 W5QFI2 A0A2R9AK31 A0A096NTB7 A0A2K6RM46 M3WK08 F7H585 A0A091DR01 A0A096NTB5 A0A2U3XFX8 A0A2U3WJR2 A0A2K6S0G9 A0A3Q0D6E4 A0A2K5URQ7 L8IU75 A0A2K6AGR0 A0A0D9R489 A0A3Q7T2L7 A5D7E4 H9ZFS7 H3AVK7 A0A2U4BC65 A0A2R9AEP9 A0A2R9ADM6 A0A2K5NC12 A0A2J8JB32 A0A3Q7SDH2 A0A2K5KC27
A0A067R9P9 A0A232EXP4 K7IQG0 A0A087ZT71 A0A2A3EDJ5 A0A0M9A6N3 E2BAW2 A0A0L7QMC6 A0A0C9QLK7 A0A158NCM0 A0A151X614 F4WFE7 A0A195F9R1 A0A195EMU4 A0A195BXI1 E2AK68 E0VSM8 E9IMC2 A0A0J7KW62 A0A1W4XP01 A0A026WA84 A0A3L8DKL5 A0A154PHF7 A0A139WFX9 A0A195CQN9 A0A1B6KBV0 A0A1S4G0D7 A0A1Y1LXY0 Q16IB0 A0A0P6J137 A0A182G802 A0A034VGH3 A0A182LS08 A0A0K8VLN3 A0A182P429 A0A0A1WJY7 A0A182QEI5 A0A182VWZ5 A0A2M4A8F1 A0A2M4A9Q0 A0A182YG12 A0A2M4A8E8 A0A0L0CC12 B0W925 A0A182RFJ9 A0A182FA37 A0A182SJS1 A0A182NDY0 W5JKE2 A0A087Z6K8 A0A2M4CVB5 A0A182K5H0 A0A182U8A4 A0A182I870 W8AM97 F5HM53 A0A384D0C3 A0A3Q7UI82 A0A3Q2H0V9 L5K2K4 A0A286ZI68 A0A182XKI2 A0A2Y9HN94 A0A2K6S0F9 Q7Q257 A0A0N8ETY7 D2H7R7 W5QFI2 A0A2R9AK31 A0A096NTB7 A0A2K6RM46 M3WK08 F7H585 A0A091DR01 A0A096NTB5 A0A2U3XFX8 A0A2U3WJR2 A0A2K6S0G9 A0A3Q0D6E4 A0A2K5URQ7 L8IU75 A0A2K6AGR0 A0A0D9R489 A0A3Q7T2L7 A5D7E4 H9ZFS7 H3AVK7 A0A2U4BC65 A0A2R9AEP9 A0A2R9ADM6 A0A2K5NC12 A0A2J8JB32 A0A3Q7SDH2 A0A2K5KC27
PDB
6QDV
E-value=0,
Score=3970
Ontologies
GO
PANTHER
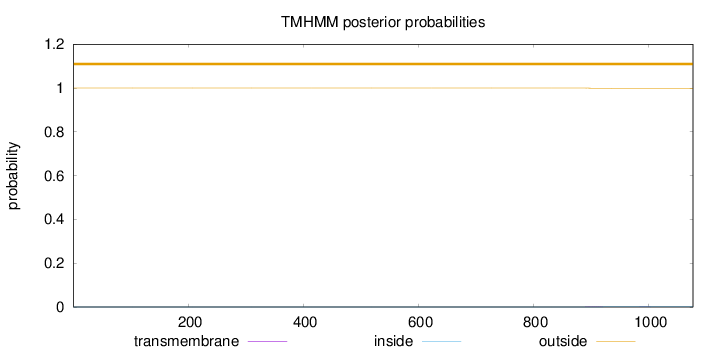
Topology
Subcellular location
Nucleus
Length:
1077
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02149
Exp number, first 60 AAs:
0.00019
Total prob of N-in:
0.00002
outside
1 - 1077
Population Genetic Test Statistics
Pi
264.891636
Theta
163.306382
Tajima's D
2.221545
CLR
0.38135
CSRT
0.91730413479326
Interpretation
Uncertain
