Gene
KWMTBOMO15205 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA004950
Annotation
PREDICTED:_C-1-tetrahydrofolate_synthase?_cytoplasmic_isoform_X2_[Bombyx_mori]
Full name
C-1-tetrahydrofolate synthase, cytoplasmic
Location in the cell
Cytoplasmic Reliability : 1.937 Mitochondrial Reliability : 1.808
Sequence
CDS
ATGTGGAAGAATTCTGAAGAATTGGAAGCTGTACAAGAGGTCACTTGCATCTTCGATCCATCTCAGTCCGAGAGAGCAGTCGGGATTGGATCGACCAACGAGACAGTTCTCTGGGCTAAGATGGCACAAATTATATCTGGGATTGAAGTTGCAGGATCTATAGAAAATGACCTGCGTCAACAAGTGACTAGGCTCCGTTCAAAATGGTCTGGCTTCGAGCCCAGGCTTGCCATCGTGCAGGTGGGCGGGCGCGAGGACTCCAACGTCTACATCAGGATGAAGCTGAAGGCAGCTGAGAAAATTGGTATCGCCGCCGAACATATCCGCTTACCAAGGGACATCACTGAAATAGAGCTGTTAGCAAAAATAACGAGTCTGAATGAATCTCCTTCGGTTCACGGCATCATTGTTCAAATGCCCCTTGATTCGGACCACGCGATCGACGCTCATCGAGTCACTGATGCTGTCTCGCCTGATAAAGATGTTGACGGCTTAAACACAATTAACGAAGGTCGAGTAGCAGTAGGTGATCTGTCCGGATTCATTCCTTGCACTCCGGCCGGGTGCGTGGAACTGATTAAGAAGACCGGCGTCACTATAGCTGGCAAAAATGTGGTAGTGCTCGGGCGGAGTCGCATTGTGGGTACTCCGGTGTCAGAGTTGCTGAAATGGGAACACGCTACTGTCACAGTGTGCCATTCAAAGACTAAGAATTTGAGTGAAATTACTAAGACAGCAGAAATACTGGTGGTGGCGATCGGTCGTCCAGAGATGGTCCGAGGTTCTTGGATCAAGCCTGGAGCGGTCGTTATAGACTGTGGTATCAACGCTATTGAGGATCCTACAAAGAAAAGCGGGCAACGTTTAGTGGGCGACGTGGCTTACCAGGAGGCGGTCAAAGTAGCTTCGGCTGTGACTCCTGTGCCGGGCGGCGTGGGGCCCATGACCGTGGCGATGCTCATGCGGAACACCGTGCAGGCAGCGCGCCGACAGCTCGACCGCTTGCTTGCCCCCTCTTGGCCTCTGAAACCGCTTCGCCTCACCCCGATTACGCCACCGCCCAGTGACATCGTAATAGCTCGTTCTCAGAAGCCTAAAGACATCGGAGAGCTGGCGGCAGAGATCGGTCTGTGTCCGAGCGAGGTGTCTCAATATGGACGTACTAAGGCCAAGATTTCACTGTCCGTTCTCGACAGACTACGCAACCAACGCACAGGGAAATACATCGTAGTTGCTGGTATAACTCCGACACCACTTGGTGAAGGTAAAAGCACCACTCTAATAGGTCTGGTGCAAGCGTTGACAGCACATCGCGGGAAGAACGCGTTTGCATGCATGCGGCAACCGAGTCAGGGACCGACCTTTGGCGTTAAAGGGGGTGCTGCTGGAGGGGGCTACTCTCAAGTAATTCCAATGGAGGAGTTCAACCTGCACATGACTGGCGACATTCATGCCGTAACCGCTGCCAACAATCTTCTTGCTGCACAAATGGACGCGCGCATCTTCCACGAGCTGACACAGAAGGACGGACCTCTGTATGACCGTCTCGTCCCCAAGATCAAAGGCGTCAGGAAGTTCTCTCCTATCCAGCTACGCAGACTGAAGAGATTGGGAATCGATAAAACGGACCCTGACTCTTTAACTGCCGAAGAGAGGGTTAAGTTCGCTCGACTCAATATAGACCCAACAAAGATTATGTGGAACAGAGTTGTGGATCTAAACGACAGATATCTTCGAAAGATTACAATAGGCCAGTCTCCTACGGAAAAAGGCTTTACTCGGGAGACTGCTTTCGATATTTCCGTCGCTTCGGAGATCATGGCTATTCTGGCGTTAGGTAATGACGTAGACGACATCAAAGAACGACTCGCCAGCATGGTGGTGGCCCTCGATAAGGACGGCAACGCTGTCACCGCAGACGACTTGGGCATGACGGGAGCGTTGCTGGTGTTGCTACGTGATGCGTTCGAGCCGACGCTGATGCAGAGTCTGGAGGGCACGCCGGTGCTGGTGCACGCAGGACCCTTCGCCAACATCGCGCACGGCTGCTCCTCCATACTGGCCGACAAGATCGCCATGAAGCTGGCCGGTCCCGCTGGATACGTCGCCACTGAAGCTGGATTTGGATCTGATATTGGAATGGAGAAGTTCTTCGACATAAAGTGTCGCGCAAGCGGTGACCGGCCTCACTGCGCGGTCATCGTTGCGACCGTGCGCGCGCTCAAGATGCACGGCGGCGGCCCGCCCGTCTCACCCGGCCTGCCTCTGCATGACGTCTACCTGCAGGAGAACCTAGATCTGCTACACAAAGGTCTATGTAACTTGGGCAAACACATCAGTAACGGCAACAAATTTGGTATTCCGGTCGTTGTAGCTGTCAACAAACACGGAAATGATAGCGCCGCGGAATTGAGTTTGGTGCAAGACTTCGCTCTGCACAACGGCGCGTTCAGGGCGGTGGTGTGTAGCCACTTCACCGACGGCGGGGCGGGCGCTATCGAGCTTGCCGACGCCGTTATCGACGCCTGCAACCGACCCACGCATTTCCAGTACCTATACCCCCTGAGCTTCTCCATCCAGGACAAGATTCACACAATCGCCAGTCAGATGTACGGCGCTTCCGAAATCGAATACACTGAAGAGGTTTTAGAAAAAATTAAAATCTTCACCGAAAAGGGATACGACAAATTTCCGATTTGCATGGCCAAGACTTCGAACTCCTTAACCGGTGATCCCTCCATCAAGGGAGCTCCCACTGACTTCACGCTAAAGATCAACGGAATATTCGTATCGGTAGGCGCCGGCTTCGTGGTGCCCATGGTCGGGGAGATCTCGAAGATGCCCGGACTGCCGACACGACCTAGCATCTACGATATTGACCTCAACACAAAAACAGGGGAAATCGAAGGCCTTTTCTAA
Protein
MWKNSEELEAVQEVTCIFDPSQSERAVGIGSTNETVLWAKMAQIISGIEVAGSIENDLRQQVTRLRSKWSGFEPRLAIVQVGGREDSNVYIRMKLKAAEKIGIAAEHIRLPRDITEIELLAKITSLNESPSVHGIIVQMPLDSDHAIDAHRVTDAVSPDKDVDGLNTINEGRVAVGDLSGFIPCTPAGCVELIKKTGVTIAGKNVVVLGRSRIVGTPVSELLKWEHATVTVCHSKTKNLSEITKTAEILVVAIGRPEMVRGSWIKPGAVVIDCGINAIEDPTKKSGQRLVGDVAYQEAVKVASAVTPVPGGVGPMTVAMLMRNTVQAARRQLDRLLAPSWPLKPLRLTPITPPPSDIVIARSQKPKDIGELAAEIGLCPSEVSQYGRTKAKISLSVLDRLRNQRTGKYIVVAGITPTPLGEGKSTTLIGLVQALTAHRGKNAFACMRQPSQGPTFGVKGGAAGGGYSQVIPMEEFNLHMTGDIHAVTAANNLLAAQMDARIFHELTQKDGPLYDRLVPKIKGVRKFSPIQLRRLKRLGIDKTDPDSLTAEERVKFARLNIDPTKIMWNRVVDLNDRYLRKITIGQSPTEKGFTRETAFDISVASEIMAILALGNDVDDIKERLASMVVALDKDGNAVTADDLGMTGALLVLLRDAFEPTLMQSLEGTPVLVHAGPFANIAHGCSSILADKIAMKLAGPAGYVATEAGFGSDIGMEKFFDIKCRASGDRPHCAVIVATVRALKMHGGGPPVSPGLPLHDVYLQENLDLLHKGLCNLGKHISNGNKFGIPVVVAVNKHGNDSAAELSLVQDFALHNGAFRAVVCSHFTDGGAGAIELADAVIDACNRPTHFQYLYPLSFSIQDKIHTIASQMYGASEIEYTEEVLEKIKIFTEKGYDKFPICMAKTSNSLTGDPSIKGAPTDFTLKINGIFVSVGAGFVVPMVGEISKMPGLPTRPSIYDIDLNTKTGEIEGLF
Summary
Catalytic Activity
(6R)-5,10-methylene-5,6,7,8-tetrahydrofolate + NADP(+) = 5,10-methenyltetrahydrofolate + NADPH
5,10-methenyltetrahydrofolate + H2O = (6S)-10-formyltetrahydrofolate + H(+)
(6S)-5,6,7,8-tetrahydrofolate + ATP + formate = (6S)-10-formyltetrahydrofolate + ADP + phosphate
5,10-methenyltetrahydrofolate + H2O = (6S)-10-formyltetrahydrofolate + H(+)
(6S)-5,6,7,8-tetrahydrofolate + ATP + formate = (6S)-10-formyltetrahydrofolate + ADP + phosphate
Subunit
Homodimer.
Similarity
In the N-terminal section; belongs to the tetrahydrofolate dehydrogenase/cyclohydrolase family.
In the C-terminal section; belongs to the formate--tetrahydrofolate ligase family.
In the C-terminal section; belongs to the formate--tetrahydrofolate ligase family.
Keywords
Amino-acid biosynthesis
ATP-binding
Cytoplasm
Histidine biosynthesis
Hydrolase
Ligase
Methionine biosynthesis
Multifunctional enzyme
NADP
Nucleotide-binding
One-carbon metabolism
Oxidoreductase
Purine biosynthesis
Alternative splicing
Complete proteome
Reference proteome
Feature
chain C-1-tetrahydrofolate synthase, cytoplasmic
splice variant In isoform A.
splice variant In isoform A.
Uniprot
H9J609
A0A1S6PVE1
A0A2H1VNX3
Q27772
A0A2A4JLQ7
A0A2W1BQ31
+ More
A0A2A4JL58 S4NT00 A0A194PG96 A0A194RQ72 A0A212F741 A0A2A4JKD0 A0A1J1J1D7 A0A1I8MEA8 A0A1I8MEB5 A0A1I8MEB1 A0A1W4WS48 A0A1Y1L059 A0A084W7B5 Q7QCW3 A0A182VRC4 A0A1S4FCI2 A0A182P257 A0A182UPM3 A0A182HTP7 A0A182XHP1 T1P9G4 A0A182RT97 A0A182QPE3 A0A1Q3FYF8 A0A1Q3FYD8 A0A182N758 A0A182J027 A0A182LWE5 A0A1Q3FYF0 W8BMC6 W8B1E2 A0A182GUE2 E9ILQ9 A0A067RJK1 A0A0M9A773 A0A0L7QK77 A0A0A1WE49 A0A182XW44 A0A0L0BV52 A0A232F5A3 A0A0C9QQT1 A0A1I8Q3K9 A0A1I8Q3M2 E2AG54 A0A034WE13 A0A034WFL5 A0A195DUB5 A0A0K8UA64 A0A1B6ED89 A0A026WV52 A0A1B6DLD3 A0A2M4BCH4 A0A1L8DZL7 A0A182F763 U5EUM8 A0A2M4CUR3 A0A2M4CTC1 A0A0A1WU59 A0A2M3ZFZ5 D6WUB5 A0A195CGD6 A0A195AY49 A0A0K8ULN7 A0A2M4CT95 A0A2A3EJF6 A0A088ABT0 A0A154P667 A0A2M4BCQ6 E0VA04 A0A2M4AEA5 A0A2M4AFE3 B4GLS8 A0A0R3NPL3 Q293Q3 A0A151XF40 A0A0R3NK86 B4PKR2 A0A0R1E6P9 A0A0R1E5B0 B3P1M0 O96553-2 E8NH74 O96553 A0A0B4K623 A0A0M5J4X3 B3LW32 B4HIS0 A0A1A9WGX4 A0A3B0K6U8 A0A336LX68 B4QV66 A0A1W4VZ59 A0A1B6G1K4 A0A1W4W008
A0A2A4JL58 S4NT00 A0A194PG96 A0A194RQ72 A0A212F741 A0A2A4JKD0 A0A1J1J1D7 A0A1I8MEA8 A0A1I8MEB5 A0A1I8MEB1 A0A1W4WS48 A0A1Y1L059 A0A084W7B5 Q7QCW3 A0A182VRC4 A0A1S4FCI2 A0A182P257 A0A182UPM3 A0A182HTP7 A0A182XHP1 T1P9G4 A0A182RT97 A0A182QPE3 A0A1Q3FYF8 A0A1Q3FYD8 A0A182N758 A0A182J027 A0A182LWE5 A0A1Q3FYF0 W8BMC6 W8B1E2 A0A182GUE2 E9ILQ9 A0A067RJK1 A0A0M9A773 A0A0L7QK77 A0A0A1WE49 A0A182XW44 A0A0L0BV52 A0A232F5A3 A0A0C9QQT1 A0A1I8Q3K9 A0A1I8Q3M2 E2AG54 A0A034WE13 A0A034WFL5 A0A195DUB5 A0A0K8UA64 A0A1B6ED89 A0A026WV52 A0A1B6DLD3 A0A2M4BCH4 A0A1L8DZL7 A0A182F763 U5EUM8 A0A2M4CUR3 A0A2M4CTC1 A0A0A1WU59 A0A2M3ZFZ5 D6WUB5 A0A195CGD6 A0A195AY49 A0A0K8ULN7 A0A2M4CT95 A0A2A3EJF6 A0A088ABT0 A0A154P667 A0A2M4BCQ6 E0VA04 A0A2M4AEA5 A0A2M4AFE3 B4GLS8 A0A0R3NPL3 Q293Q3 A0A151XF40 A0A0R3NK86 B4PKR2 A0A0R1E6P9 A0A0R1E5B0 B3P1M0 O96553-2 E8NH74 O96553 A0A0B4K623 A0A0M5J4X3 B3LW32 B4HIS0 A0A1A9WGX4 A0A3B0K6U8 A0A336LX68 B4QV66 A0A1W4VZ59 A0A1B6G1K4 A0A1W4W008
Pubmed
19121390
7893749
28756777
23622113
26354079
22118469
+ More
25315136 28004739 24438588 12364791 14747013 17210077 17510324 24495485 26483478 21282665 24845553 25830018 25244985 26108605 28648823 20798317 25348373 24508170 30249741 18362917 19820115 20566863 17994087 15632085 17550304 9832531 10731132 12537572 12537569 12537568 12537573 12537574 16110336 17569856 17569867
25315136 28004739 24438588 12364791 14747013 17210077 17510324 24495485 26483478 21282665 24845553 25830018 25244985 26108605 28648823 20798317 25348373 24508170 30249741 18362917 19820115 20566863 17994087 15632085 17550304 9832531 10731132 12537572 12537569 12537568 12537573 12537574 16110336 17569856 17569867
EMBL
BABH01029887
KY622019
AQV04396.1
ODYU01003584
SOQ42530.1
L36189
+ More
NWSH01001119 PCG72504.1 KZ150014 PZC74996.1 PCG72506.1 GAIX01013842 JAA78718.1 KQ459604 KPI92322.1 KQ460045 KPJ18171.1 AGBW02009936 OWR49547.1 PCG72507.1 CVRI01000066 CRL06152.1 GEZM01071446 JAV65800.1 ATLV01021193 ATLV01021194 ATLV01021195 KE525314 KFB46109.1 AAAB01008859 EAA07766.5 APCN01000574 KA645349 AFP59978.1 AXCN02000326 GFDL01002495 JAV32550.1 GFDL01002497 JAV32548.1 AXCM01002203 GFDL01002487 JAV32558.1 GAMC01015725 GAMC01015721 JAB90834.1 GAMC01015722 GAMC01015720 JAB90833.1 JXUM01088996 JXUM01088997 JXUM01088998 JXUM01088999 KQ563739 KXJ73402.1 GL764129 EFZ18327.1 KK852643 KDR19573.1 KQ435733 KOX77269.1 KQ414983 KOC59022.1 GBXI01017018 JAC97273.1 JRES01001414 KNC23124.1 NNAY01000972 OXU25660.1 GBYB01003027 JAG72794.1 GL439266 EFN67545.1 GAKP01006023 JAC52929.1 GAKP01006022 JAC52930.1 KQ980419 KYN16129.1 GDHF01028770 JAI23544.1 GEDC01001389 JAS35909.1 KK107087 QOIP01000001 EZA59897.1 RLU27323.1 GEDC01010846 JAS26452.1 GGFJ01001613 MBW50754.1 GFDF01002217 JAV11867.1 GANO01002248 JAB57623.1 GGFL01004400 MBW68578.1 GGFL01004399 MBW68577.1 GBXI01012121 JAD02171.1 GGFM01006723 MBW27474.1 KQ971352 EFA07236.1 KQ977791 KYM99782.1 KQ976703 KYM77173.1 GDHF01024843 JAI27471.1 GGFL01004398 MBW68576.1 KZ288239 PBC31316.1 KQ434809 KZC06698.1 GGFJ01001661 MBW50802.1 DS235004 EEB10210.1 GGFK01005796 MBW39117.1 GGFK01006162 MBW39483.1 CH479185 EDW38502.1 CM000070 KRT01327.1 EAL29161.2 KQ982215 KYQ58888.1 KRT01326.1 CM000160 EDW96821.1 KRK03378.1 KRK03379.1 CH954181 EDV49619.1 AF082097 AE014297 BT021278 AY069146 BT125950 ADX35929.1 AFH06346.1 CP012526 ALC46524.1 CH902617 EDV41565.1 CH480815 EDW42717.1 OUUW01000008 SPP83790.1 UFQT01000188 SSX21369.1 CM000364 EDX13472.1 GECZ01013459 JAS56310.1
NWSH01001119 PCG72504.1 KZ150014 PZC74996.1 PCG72506.1 GAIX01013842 JAA78718.1 KQ459604 KPI92322.1 KQ460045 KPJ18171.1 AGBW02009936 OWR49547.1 PCG72507.1 CVRI01000066 CRL06152.1 GEZM01071446 JAV65800.1 ATLV01021193 ATLV01021194 ATLV01021195 KE525314 KFB46109.1 AAAB01008859 EAA07766.5 APCN01000574 KA645349 AFP59978.1 AXCN02000326 GFDL01002495 JAV32550.1 GFDL01002497 JAV32548.1 AXCM01002203 GFDL01002487 JAV32558.1 GAMC01015725 GAMC01015721 JAB90834.1 GAMC01015722 GAMC01015720 JAB90833.1 JXUM01088996 JXUM01088997 JXUM01088998 JXUM01088999 KQ563739 KXJ73402.1 GL764129 EFZ18327.1 KK852643 KDR19573.1 KQ435733 KOX77269.1 KQ414983 KOC59022.1 GBXI01017018 JAC97273.1 JRES01001414 KNC23124.1 NNAY01000972 OXU25660.1 GBYB01003027 JAG72794.1 GL439266 EFN67545.1 GAKP01006023 JAC52929.1 GAKP01006022 JAC52930.1 KQ980419 KYN16129.1 GDHF01028770 JAI23544.1 GEDC01001389 JAS35909.1 KK107087 QOIP01000001 EZA59897.1 RLU27323.1 GEDC01010846 JAS26452.1 GGFJ01001613 MBW50754.1 GFDF01002217 JAV11867.1 GANO01002248 JAB57623.1 GGFL01004400 MBW68578.1 GGFL01004399 MBW68577.1 GBXI01012121 JAD02171.1 GGFM01006723 MBW27474.1 KQ971352 EFA07236.1 KQ977791 KYM99782.1 KQ976703 KYM77173.1 GDHF01024843 JAI27471.1 GGFL01004398 MBW68576.1 KZ288239 PBC31316.1 KQ434809 KZC06698.1 GGFJ01001661 MBW50802.1 DS235004 EEB10210.1 GGFK01005796 MBW39117.1 GGFK01006162 MBW39483.1 CH479185 EDW38502.1 CM000070 KRT01327.1 EAL29161.2 KQ982215 KYQ58888.1 KRT01326.1 CM000160 EDW96821.1 KRK03378.1 KRK03379.1 CH954181 EDV49619.1 AF082097 AE014297 BT021278 AY069146 BT125950 ADX35929.1 AFH06346.1 CP012526 ALC46524.1 CH902617 EDV41565.1 CH480815 EDW42717.1 OUUW01000008 SPP83790.1 UFQT01000188 SSX21369.1 CM000364 EDX13472.1 GECZ01013459 JAS56310.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000183832
+ More
UP000095301 UP000192223 UP000030765 UP000007062 UP000075920 UP000075885 UP000075903 UP000075840 UP000076407 UP000075900 UP000075886 UP000075884 UP000075880 UP000075883 UP000069940 UP000249989 UP000027135 UP000053105 UP000053825 UP000076408 UP000037069 UP000215335 UP000095300 UP000000311 UP000078492 UP000053097 UP000279307 UP000069272 UP000007266 UP000078542 UP000078540 UP000242457 UP000005203 UP000076502 UP000009046 UP000008744 UP000001819 UP000075809 UP000002282 UP000008711 UP000000803 UP000092553 UP000007801 UP000001292 UP000091820 UP000268350 UP000000304 UP000192221
UP000095301 UP000192223 UP000030765 UP000007062 UP000075920 UP000075885 UP000075903 UP000075840 UP000076407 UP000075900 UP000075886 UP000075884 UP000075880 UP000075883 UP000069940 UP000249989 UP000027135 UP000053105 UP000053825 UP000076408 UP000037069 UP000215335 UP000095300 UP000000311 UP000078492 UP000053097 UP000279307 UP000069272 UP000007266 UP000078542 UP000078540 UP000242457 UP000005203 UP000076502 UP000009046 UP000008744 UP000001819 UP000075809 UP000002282 UP000008711 UP000000803 UP000092553 UP000007801 UP000001292 UP000091820 UP000268350 UP000000304 UP000192221
Interpro
IPR036291
NAD(P)-bd_dom_sf
+ More
IPR020630 THF_DH/CycHdrlase_cat_dom
IPR000672 THF_DH/CycHdrlase
IPR020628 Formate_THF_ligase_CS
IPR027417 P-loop_NTPase
IPR020867 THF_DH/CycHdrlase_CS
IPR000559 Formate_THF_ligase
IPR020631 THF_DH/CycHdrlase_NAD-bd_dom
IPR002903 RsmH
IPR023397 SAM-dep_MeTrfase_MraW_recog
IPR029063 SAM-dependent_MTases
IPR020630 THF_DH/CycHdrlase_cat_dom
IPR000672 THF_DH/CycHdrlase
IPR020628 Formate_THF_ligase_CS
IPR027417 P-loop_NTPase
IPR020867 THF_DH/CycHdrlase_CS
IPR000559 Formate_THF_ligase
IPR020631 THF_DH/CycHdrlase_NAD-bd_dom
IPR002903 RsmH
IPR023397 SAM-dep_MeTrfase_MraW_recog
IPR029063 SAM-dependent_MTases
ProteinModelPortal
H9J609
A0A1S6PVE1
A0A2H1VNX3
Q27772
A0A2A4JLQ7
A0A2W1BQ31
+ More
A0A2A4JL58 S4NT00 A0A194PG96 A0A194RQ72 A0A212F741 A0A2A4JKD0 A0A1J1J1D7 A0A1I8MEA8 A0A1I8MEB5 A0A1I8MEB1 A0A1W4WS48 A0A1Y1L059 A0A084W7B5 Q7QCW3 A0A182VRC4 A0A1S4FCI2 A0A182P257 A0A182UPM3 A0A182HTP7 A0A182XHP1 T1P9G4 A0A182RT97 A0A182QPE3 A0A1Q3FYF8 A0A1Q3FYD8 A0A182N758 A0A182J027 A0A182LWE5 A0A1Q3FYF0 W8BMC6 W8B1E2 A0A182GUE2 E9ILQ9 A0A067RJK1 A0A0M9A773 A0A0L7QK77 A0A0A1WE49 A0A182XW44 A0A0L0BV52 A0A232F5A3 A0A0C9QQT1 A0A1I8Q3K9 A0A1I8Q3M2 E2AG54 A0A034WE13 A0A034WFL5 A0A195DUB5 A0A0K8UA64 A0A1B6ED89 A0A026WV52 A0A1B6DLD3 A0A2M4BCH4 A0A1L8DZL7 A0A182F763 U5EUM8 A0A2M4CUR3 A0A2M4CTC1 A0A0A1WU59 A0A2M3ZFZ5 D6WUB5 A0A195CGD6 A0A195AY49 A0A0K8ULN7 A0A2M4CT95 A0A2A3EJF6 A0A088ABT0 A0A154P667 A0A2M4BCQ6 E0VA04 A0A2M4AEA5 A0A2M4AFE3 B4GLS8 A0A0R3NPL3 Q293Q3 A0A151XF40 A0A0R3NK86 B4PKR2 A0A0R1E6P9 A0A0R1E5B0 B3P1M0 O96553-2 E8NH74 O96553 A0A0B4K623 A0A0M5J4X3 B3LW32 B4HIS0 A0A1A9WGX4 A0A3B0K6U8 A0A336LX68 B4QV66 A0A1W4VZ59 A0A1B6G1K4 A0A1W4W008
A0A2A4JL58 S4NT00 A0A194PG96 A0A194RQ72 A0A212F741 A0A2A4JKD0 A0A1J1J1D7 A0A1I8MEA8 A0A1I8MEB5 A0A1I8MEB1 A0A1W4WS48 A0A1Y1L059 A0A084W7B5 Q7QCW3 A0A182VRC4 A0A1S4FCI2 A0A182P257 A0A182UPM3 A0A182HTP7 A0A182XHP1 T1P9G4 A0A182RT97 A0A182QPE3 A0A1Q3FYF8 A0A1Q3FYD8 A0A182N758 A0A182J027 A0A182LWE5 A0A1Q3FYF0 W8BMC6 W8B1E2 A0A182GUE2 E9ILQ9 A0A067RJK1 A0A0M9A773 A0A0L7QK77 A0A0A1WE49 A0A182XW44 A0A0L0BV52 A0A232F5A3 A0A0C9QQT1 A0A1I8Q3K9 A0A1I8Q3M2 E2AG54 A0A034WE13 A0A034WFL5 A0A195DUB5 A0A0K8UA64 A0A1B6ED89 A0A026WV52 A0A1B6DLD3 A0A2M4BCH4 A0A1L8DZL7 A0A182F763 U5EUM8 A0A2M4CUR3 A0A2M4CTC1 A0A0A1WU59 A0A2M3ZFZ5 D6WUB5 A0A195CGD6 A0A195AY49 A0A0K8ULN7 A0A2M4CT95 A0A2A3EJF6 A0A088ABT0 A0A154P667 A0A2M4BCQ6 E0VA04 A0A2M4AEA5 A0A2M4AFE3 B4GLS8 A0A0R3NPL3 Q293Q3 A0A151XF40 A0A0R3NK86 B4PKR2 A0A0R1E6P9 A0A0R1E5B0 B3P1M0 O96553-2 E8NH74 O96553 A0A0B4K623 A0A0M5J4X3 B3LW32 B4HIS0 A0A1A9WGX4 A0A3B0K6U8 A0A336LX68 B4QV66 A0A1W4VZ59 A0A1B6G1K4 A0A1W4W008
PDB
4JKI
E-value=2.34062e-139,
Score=1274
Ontologies
PATHWAY
GO
Topology
Subcellular location
Cytoplasm
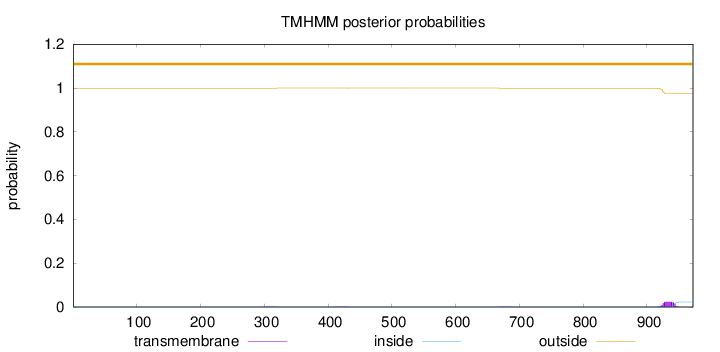
Length:
972
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.50644
Exp number, first 60 AAs:
0.00109
Total prob of N-in:
0.00055
outside
1 - 972
Population Genetic Test Statistics
Pi
271.465554
Theta
185.604193
Tajima's D
1.752803
CLR
0.185425
CSRT
0.84120793960302
Interpretation
Uncertain
