Gene
KWMTBOMO15201
Pre Gene Modal
BGIBMGA004964
Annotation
PREDICTED:_transcriptional_adapter_2B_isoform_X1_[Bombyx_mori]
Full name
Transcriptional adapter
+ More
Transcriptional adapter 2B
Transcriptional adapter 2B
Alternative Name
dADA2b
Transcription factor
Location in the cell
Nuclear Reliability : 3.602
Sequence
CDS
ATGTCATTTTCAGATCTTTACGCTAAATATAATTGTACATATTGTCAAGAGGAAATAAACGGCGTAAGAGTTAAATGTACTGAATGTGTTGATTTTGATATTTGTTTGCAGTGTTTTTCTCTTGGTGCTGAAATCGGACAGCACAAAAACGATCATTCATATCAGTTTGTGGACTCTGGGGCTTTTGGAATATTTTTAGGACGTAGTAGCTGGTCAGCTAACGAAGAAGTAAGATTATTAGATGCCATAGAACAATTCGGCTTTGGTAACTGGGAGGATATAGCAAAACATATTGAAACAAGAACTCCTGAAGAAGCAAAAGATGAATACATTACTAGGTACCTGGAAGGCACCATAGGCAAAGCAACATGGGGCAATATTGAAAGTACATCTCGCCCATCACTACATTGTGCAGATAAAGATGATGGCCCACTTGGTCCAACTGCAGTGTCTCGACTACCAAACCTAGCTATAACAGTAGACGAAGCAGCACAACTTGGGTATATGCCAAATAGAGATGATTTTGAAAGAGAACATGATCATGAGGCAGAGCAGTTGATCTCAACATTATCATTAAATCCAGAAGATGATGAATTAGATGTTGCACTAAAGCTATCACAAGTTGATATATACATCAGAAGACTGAGAGAGAGAACGAGACGAAAAAGATTAGTTAGAGATTTCCAGTTAGTGTCTGTCTTCTTCAACAATCAGAGGAATAAACAGAAGACCCTAGGAAAACTTGCCAAAGAAAAAAAAGAGTTCACGGAGCGGCTACGATGGATGGCCCAATTCTACGGGCGCGCTGAGCAGGCGTGCGTGTCGAGCGGGCTGTGGCGGGAGCGCGAGCTGCGCGTGCGGCTGGCGGAGCTGCACCGGTACCGGCTGGCCGGCGTCACGCGGCTGGAGGAGTGCGCGCACCACGACCAGCACGCCGCGCTCAGGCGCTCCACGCACCACGACCGGAACGGTAGCAGTGGGTGCCCGGAAACCAATCAGACAAGAGATCAAACATCGATCAACTCGCTGCAGCAAAGAAAAAGAGACGGCGAAAGCGGCTCAAATTCCACCAGCCCAAAGTGCACACGGGACGGAAGTACCGCATGTGGATGCTCCAAAAAGAGCTCATGCAACAGCAACAGCGCAGGATGCTCGAGGAATCTGCTCACCACTAATGAGATACAGTTGTGTTCGGCGTTGAGCCTGCCGCCGGCGCAGTACGTGACGCTGAAGGGCGTGTTGCTCCGCCGAGCGCCCGCGCCCGACTGCGACCTCGACCTCTCCGTCACGCACTACTTGCACGCCGCCGGATGGATACGGAACTAG
Protein
MSFSDLYAKYNCTYCQEEINGVRVKCTECVDFDICLQCFSLGAEIGQHKNDHSYQFVDSGAFGIFLGRSSWSANEEVRLLDAIEQFGFGNWEDIAKHIETRTPEEAKDEYITRYLEGTIGKATWGNIESTSRPSLHCADKDDGPLGPTAVSRLPNLAITVDEAAQLGYMPNRDDFEREHDHEAEQLISTLSLNPEDDELDVALKLSQVDIYIRRLRERTRRKRLVRDFQLVSVFFNNQRNKQKTLGKLAKEKKEFTERLRWMAQFYGRAEQACVSSGLWRERELRVRLAELHRYRLAGVTRLEECAHHDQHAALRRSTHHDRNGSSGCPETNQTRDQTSINSLQQRKRDGESGSNSTSPKCTRDGSTACGCSKKSSCNSNSAGCSRNLLTTNEIQLCSALSLPPAQYVTLKGVLLRRAPAPDCDLDLSVTHYLHAAGWIRN
Summary
Description
Required for the function of some acidic activation domains, which activate transcription from a distant site. Binds double-stranded DNA. Binds dinucleosomes, probably at the linker region between neighboring nucleosomes. Plays a role in chromatin remodeling.
Subunit
Interacts with Gcn5 and Ada3 as part of a GCN5-containing ADA multiprotein complex (PubMed:12482983). Associated with a SAGA-type complex that also contains Taf5, e(y)1/Taf9, Taf10, Spt3 and Taf1 (PubMed:12697829).
Keywords
Alternative splicing
Complete proteome
DNA-binding
Metal-binding
Nucleus
Reference proteome
Transcription
Transcription regulation
Zinc
Zinc-finger
Feature
chain Transcriptional adapter 2B
splice variant In isoform A.
splice variant In isoform A.
Uniprot
A0A2A4JG19
A0A212F744
A0A194PGZ6
A0A194RL76
H9J623
A0A2A4JGD2
+ More
A0A067RT11 A0A2J7R553 K7J1E2 A0A154P6B1 A0A232F1X3 A0A182H092 A0A0C9RTN3 A0A0C9RBJ0 A0A310SKC9 A0A0C9RCQ9 A0A2A3EPN6 A0A088AL74 E2CAG1 Q17J17 A0A026VVT0 E2ACN3 A0A151X002 A0A151IQP3 B0WJF7 D6WTS8 A0A1Y1KVP5 A0A139WFG5 Q8I8V0 A0A1W4XDN2 A0A0K8TLB3 A0A023ENK0 B4QYP2 Q8I8V0-2 A0A0B4KFU2 B4HLP3 A0A1I8NBA0 I5ANT8 A0A1I8NBB5 A0A0Q9X563 B4PSM1 A0A0R1E670 A0A0P9ATK8 Q299A8 A0A182MN65 A0A0R3NMD7 B3LZW9 A0A0P8Y1L2 B4NK22 A0A3B0JQ79 A0A1W4W1W8 A0A1B0CMT4 B3NZW3 A0A2M4AWM1 B4JF49 A0A1I8NBA3 Q17J15 Q17J16 A0A2C9GQG4 A0A1W4VQD1 A0A182VDC6 A0A1I8JW23 Q7PQQ9 W8CDD8 A0A3B0JRR0 A0A1L8DX02 W5JW43 A0A182LC52 A0A1L8DWX3 A0A0N0BI82 A0A182R096 A0A0Q9WT02 A0A1I8JW03 F5HKZ2 A0A182TXI8 A0A2M4AUK8 B4M162 A0A2M4AUJ3 W8C9J1 A0A1Q3F7G0 B4K961 A0A1Q3G2E5 A0A2P8YRH3 A0A034VK62 A0A0K8W8L7 A0A1Y1KQ99 A0A0A1WSH4 A0A0K8VB12 A0A1A9YQ64 A0A1B0B2Z1 A0A1A9VDP4 A0A034VK74 A0A1I8NTY5 A0A0K8WF24 A0A0A1WU88 A0A1A9W5P1 A0A1I8NTY2 A0A1Y1KPX8 A0A034VHW0
A0A067RT11 A0A2J7R553 K7J1E2 A0A154P6B1 A0A232F1X3 A0A182H092 A0A0C9RTN3 A0A0C9RBJ0 A0A310SKC9 A0A0C9RCQ9 A0A2A3EPN6 A0A088AL74 E2CAG1 Q17J17 A0A026VVT0 E2ACN3 A0A151X002 A0A151IQP3 B0WJF7 D6WTS8 A0A1Y1KVP5 A0A139WFG5 Q8I8V0 A0A1W4XDN2 A0A0K8TLB3 A0A023ENK0 B4QYP2 Q8I8V0-2 A0A0B4KFU2 B4HLP3 A0A1I8NBA0 I5ANT8 A0A1I8NBB5 A0A0Q9X563 B4PSM1 A0A0R1E670 A0A0P9ATK8 Q299A8 A0A182MN65 A0A0R3NMD7 B3LZW9 A0A0P8Y1L2 B4NK22 A0A3B0JQ79 A0A1W4W1W8 A0A1B0CMT4 B3NZW3 A0A2M4AWM1 B4JF49 A0A1I8NBA3 Q17J15 Q17J16 A0A2C9GQG4 A0A1W4VQD1 A0A182VDC6 A0A1I8JW23 Q7PQQ9 W8CDD8 A0A3B0JRR0 A0A1L8DX02 W5JW43 A0A182LC52 A0A1L8DWX3 A0A0N0BI82 A0A182R096 A0A0Q9WT02 A0A1I8JW03 F5HKZ2 A0A182TXI8 A0A2M4AUK8 B4M162 A0A2M4AUJ3 W8C9J1 A0A1Q3F7G0 B4K961 A0A1Q3G2E5 A0A2P8YRH3 A0A034VK62 A0A0K8W8L7 A0A1Y1KQ99 A0A0A1WSH4 A0A0K8VB12 A0A1A9YQ64 A0A1B0B2Z1 A0A1A9VDP4 A0A034VK74 A0A1I8NTY5 A0A0K8WF24 A0A0A1WU88 A0A1A9W5P1 A0A1I8NTY2 A0A1Y1KPX8 A0A034VHW0
Pubmed
22118469
26354079
19121390
24845553
20075255
28648823
+ More
26483478 20798317 17510324 24508170 18362917 19820115 28004739 12697829 10731132 12537572 12537569 12482983 26369729 24945155 17994087 12537568 12537573 12537574 16110336 17569856 17569867 25315136 15632085 17550304 12364791 14747013 17210077 24495485 20920257 23761445 20966253 29403074 25348373 25830018
26483478 20798317 17510324 24508170 18362917 19820115 28004739 12697829 10731132 12537572 12537569 12482983 26369729 24945155 17994087 12537568 12537573 12537574 16110336 17569856 17569867 25315136 15632085 17550304 12364791 14747013 17210077 24495485 20920257 23761445 20966253 29403074 25348373 25830018
EMBL
NWSH01001593
PCG70769.1
AGBW02009936
OWR49544.1
KQ459604
KPI92318.1
+ More
KQ460045 KPJ18174.1 BABH01029890 PCG70768.1 KK852433 KDR23950.1 NEVH01007395 PNF35964.1 KQ434809 KZC06680.1 NNAY01001210 OXU24725.1 JXUM01100897 KQ564607 KXJ72032.1 GBYB01010911 JAG80678.1 GBYB01004251 JAG74018.1 KQ763472 OAD55041.1 GBYB01010907 JAG80674.1 KZ288209 PBC33001.1 GL453976 EFN75072.1 CH477235 EAT46696.1 KK107790 EZA47770.1 GL438568 EFN68777.1 KQ982625 KYQ53469.1 KQ976755 KYN08562.1 DS231959 EDS29131.1 KQ971352 EFA06279.1 GEZM01077000 JAV63476.1 KYB26672.1 AY142213 AE014297 AY058546 AAL13775.1 AAN52141.1 AAZ52519.1 GDAI01002466 JAI15137.1 GAPW01002606 JAC10992.1 CM000364 EDX13800.1 AGB95749.1 CH480815 EDW43070.1 CM000070 EIM52623.1 CH964272 KRG00178.1 CM000160 EDW96481.1 KRK03172.1 CH902617 KPU80642.1 EAL27795.2 AXCM01000265 KRT00291.1 EDV44159.1 KPU80643.1 EDW85064.1 OUUW01000007 SPP83073.1 AJWK01019156 AJWK01019157 CH954181 EDV49961.1 GGFK01011876 MBW45197.1 CH916369 EDV93330.1 EAT46695.1 EAT46694.1 APCN01002106 AAAB01008879 EAA08449.6 GAMC01002121 JAC04435.1 SPP83072.1 GFDF01003116 JAV10968.1 ADMH02000272 ETN67169.1 GFDF01003117 JAV10967.1 KQ435733 KOX77288.1 AXCN02000226 CH940650 KRF83301.1 EGK96953.1 GGFK01011144 MBW44465.1 EDW67473.1 GGFK01011129 MBW44450.1 GAMC01002119 JAC04437.1 GFDL01011541 JAV23504.1 CH933806 EDW14474.1 GFDL01001073 JAV33972.1 PYGN01000411 PSN46846.1 GAKP01016073 JAC42879.1 GDHF01004892 JAI47422.1 GEZM01077003 JAV63474.1 GBXI01012696 JAD01596.1 GDHF01016246 JAI36068.1 JXJN01007738 GAKP01016058 JAC42894.1 GDHF01002660 JAI49654.1 GBXI01012304 JAD01988.1 GEZM01077005 GEZM01077004 JAV63473.1 GAKP01016056 JAC42896.1
KQ460045 KPJ18174.1 BABH01029890 PCG70768.1 KK852433 KDR23950.1 NEVH01007395 PNF35964.1 KQ434809 KZC06680.1 NNAY01001210 OXU24725.1 JXUM01100897 KQ564607 KXJ72032.1 GBYB01010911 JAG80678.1 GBYB01004251 JAG74018.1 KQ763472 OAD55041.1 GBYB01010907 JAG80674.1 KZ288209 PBC33001.1 GL453976 EFN75072.1 CH477235 EAT46696.1 KK107790 EZA47770.1 GL438568 EFN68777.1 KQ982625 KYQ53469.1 KQ976755 KYN08562.1 DS231959 EDS29131.1 KQ971352 EFA06279.1 GEZM01077000 JAV63476.1 KYB26672.1 AY142213 AE014297 AY058546 AAL13775.1 AAN52141.1 AAZ52519.1 GDAI01002466 JAI15137.1 GAPW01002606 JAC10992.1 CM000364 EDX13800.1 AGB95749.1 CH480815 EDW43070.1 CM000070 EIM52623.1 CH964272 KRG00178.1 CM000160 EDW96481.1 KRK03172.1 CH902617 KPU80642.1 EAL27795.2 AXCM01000265 KRT00291.1 EDV44159.1 KPU80643.1 EDW85064.1 OUUW01000007 SPP83073.1 AJWK01019156 AJWK01019157 CH954181 EDV49961.1 GGFK01011876 MBW45197.1 CH916369 EDV93330.1 EAT46695.1 EAT46694.1 APCN01002106 AAAB01008879 EAA08449.6 GAMC01002121 JAC04435.1 SPP83072.1 GFDF01003116 JAV10968.1 ADMH02000272 ETN67169.1 GFDF01003117 JAV10967.1 KQ435733 KOX77288.1 AXCN02000226 CH940650 KRF83301.1 EGK96953.1 GGFK01011144 MBW44465.1 EDW67473.1 GGFK01011129 MBW44450.1 GAMC01002119 JAC04437.1 GFDL01011541 JAV23504.1 CH933806 EDW14474.1 GFDL01001073 JAV33972.1 PYGN01000411 PSN46846.1 GAKP01016073 JAC42879.1 GDHF01004892 JAI47422.1 GEZM01077003 JAV63474.1 GBXI01012696 JAD01596.1 GDHF01016246 JAI36068.1 JXJN01007738 GAKP01016058 JAC42894.1 GDHF01002660 JAI49654.1 GBXI01012304 JAD01988.1 GEZM01077005 GEZM01077004 JAV63473.1 GAKP01016056 JAC42896.1
Proteomes
UP000218220
UP000007151
UP000053268
UP000053240
UP000005204
UP000027135
+ More
UP000235965 UP000002358 UP000076502 UP000215335 UP000069940 UP000249989 UP000242457 UP000005203 UP000008237 UP000008820 UP000053097 UP000000311 UP000075809 UP000078542 UP000002320 UP000007266 UP000000803 UP000192223 UP000000304 UP000001292 UP000095301 UP000001819 UP000007798 UP000002282 UP000007801 UP000075883 UP000268350 UP000192221 UP000092461 UP000008711 UP000001070 UP000075840 UP000075903 UP000076407 UP000007062 UP000000673 UP000075882 UP000053105 UP000075886 UP000008792 UP000075902 UP000009192 UP000245037 UP000092443 UP000092460 UP000078200 UP000095300 UP000091820
UP000235965 UP000002358 UP000076502 UP000215335 UP000069940 UP000249989 UP000242457 UP000005203 UP000008237 UP000008820 UP000053097 UP000000311 UP000075809 UP000078542 UP000002320 UP000007266 UP000000803 UP000192223 UP000000304 UP000001292 UP000095301 UP000001819 UP000007798 UP000002282 UP000007801 UP000075883 UP000268350 UP000192221 UP000092461 UP000008711 UP000001070 UP000075840 UP000075903 UP000076407 UP000007062 UP000000673 UP000075882 UP000053105 UP000075886 UP000008792 UP000075902 UP000009192 UP000245037 UP000092443 UP000092460 UP000078200 UP000095300 UP000091820
Interpro
Gene 3D
ProteinModelPortal
A0A2A4JG19
A0A212F744
A0A194PGZ6
A0A194RL76
H9J623
A0A2A4JGD2
+ More
A0A067RT11 A0A2J7R553 K7J1E2 A0A154P6B1 A0A232F1X3 A0A182H092 A0A0C9RTN3 A0A0C9RBJ0 A0A310SKC9 A0A0C9RCQ9 A0A2A3EPN6 A0A088AL74 E2CAG1 Q17J17 A0A026VVT0 E2ACN3 A0A151X002 A0A151IQP3 B0WJF7 D6WTS8 A0A1Y1KVP5 A0A139WFG5 Q8I8V0 A0A1W4XDN2 A0A0K8TLB3 A0A023ENK0 B4QYP2 Q8I8V0-2 A0A0B4KFU2 B4HLP3 A0A1I8NBA0 I5ANT8 A0A1I8NBB5 A0A0Q9X563 B4PSM1 A0A0R1E670 A0A0P9ATK8 Q299A8 A0A182MN65 A0A0R3NMD7 B3LZW9 A0A0P8Y1L2 B4NK22 A0A3B0JQ79 A0A1W4W1W8 A0A1B0CMT4 B3NZW3 A0A2M4AWM1 B4JF49 A0A1I8NBA3 Q17J15 Q17J16 A0A2C9GQG4 A0A1W4VQD1 A0A182VDC6 A0A1I8JW23 Q7PQQ9 W8CDD8 A0A3B0JRR0 A0A1L8DX02 W5JW43 A0A182LC52 A0A1L8DWX3 A0A0N0BI82 A0A182R096 A0A0Q9WT02 A0A1I8JW03 F5HKZ2 A0A182TXI8 A0A2M4AUK8 B4M162 A0A2M4AUJ3 W8C9J1 A0A1Q3F7G0 B4K961 A0A1Q3G2E5 A0A2P8YRH3 A0A034VK62 A0A0K8W8L7 A0A1Y1KQ99 A0A0A1WSH4 A0A0K8VB12 A0A1A9YQ64 A0A1B0B2Z1 A0A1A9VDP4 A0A034VK74 A0A1I8NTY5 A0A0K8WF24 A0A0A1WU88 A0A1A9W5P1 A0A1I8NTY2 A0A1Y1KPX8 A0A034VHW0
A0A067RT11 A0A2J7R553 K7J1E2 A0A154P6B1 A0A232F1X3 A0A182H092 A0A0C9RTN3 A0A0C9RBJ0 A0A310SKC9 A0A0C9RCQ9 A0A2A3EPN6 A0A088AL74 E2CAG1 Q17J17 A0A026VVT0 E2ACN3 A0A151X002 A0A151IQP3 B0WJF7 D6WTS8 A0A1Y1KVP5 A0A139WFG5 Q8I8V0 A0A1W4XDN2 A0A0K8TLB3 A0A023ENK0 B4QYP2 Q8I8V0-2 A0A0B4KFU2 B4HLP3 A0A1I8NBA0 I5ANT8 A0A1I8NBB5 A0A0Q9X563 B4PSM1 A0A0R1E670 A0A0P9ATK8 Q299A8 A0A182MN65 A0A0R3NMD7 B3LZW9 A0A0P8Y1L2 B4NK22 A0A3B0JQ79 A0A1W4W1W8 A0A1B0CMT4 B3NZW3 A0A2M4AWM1 B4JF49 A0A1I8NBA3 Q17J15 Q17J16 A0A2C9GQG4 A0A1W4VQD1 A0A182VDC6 A0A1I8JW23 Q7PQQ9 W8CDD8 A0A3B0JRR0 A0A1L8DX02 W5JW43 A0A182LC52 A0A1L8DWX3 A0A0N0BI82 A0A182R096 A0A0Q9WT02 A0A1I8JW03 F5HKZ2 A0A182TXI8 A0A2M4AUK8 B4M162 A0A2M4AUJ3 W8C9J1 A0A1Q3F7G0 B4K961 A0A1Q3G2E5 A0A2P8YRH3 A0A034VK62 A0A0K8W8L7 A0A1Y1KQ99 A0A0A1WSH4 A0A0K8VB12 A0A1A9YQ64 A0A1B0B2Z1 A0A1A9VDP4 A0A034VK74 A0A1I8NTY5 A0A0K8WF24 A0A0A1WU88 A0A1A9W5P1 A0A1I8NTY2 A0A1Y1KPX8 A0A034VHW0
PDB
6CW2
E-value=1.45898e-19,
Score=237
Ontologies
GO
Topology
Subcellular location
Nucleus
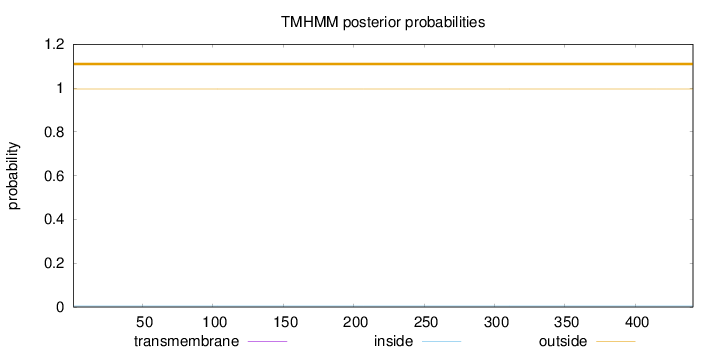
Length:
441
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.000940000000000001
Exp number, first 60 AAs:
0.0002
Total prob of N-in:
0.00431
outside
1 - 441
Population Genetic Test Statistics
Pi
284.425687
Theta
188.853458
Tajima's D
1.594244
CLR
0.00581
CSRT
0.80265986700665
Interpretation
Uncertain
