Gene
KWMTBOMO15199
Pre Gene Modal
BGIBMGA004946
Annotation
PREDICTED:_cyclin-dependent_kinase_20_[Papilio_xuthus]
Full name
Cyclin-dependent kinase 20
Alternative Name
Cell cycle-related kinase
Cell division protein kinase 20
CDK-activating kinase p42
CDK-related protein kinase PNQLARE
Cyclin-dependent protein kinase H
Cyclin-kinase-activating kinase p42
Cell division protein kinase 20
CDK-activating kinase p42
CDK-related protein kinase PNQLARE
Cyclin-dependent protein kinase H
Cyclin-kinase-activating kinase p42
Location in the cell
Cytoplasmic Reliability : 1.509 Mitochondrial Reliability : 2.048
Sequence
CDS
ATGGATTATGATGTTAATAATTATACAGTCGTTGGTCGAATCGGTGAAGGAGCCCACGGGCTTGTTTTCAAAGCCAAACACAACACAACAGGTCGCGAGGTGGCCTTAAAGAAAATTCTGATAAAGAATCTTGAAGATGGAATACCGGTCCATGTCATGAGAGAGATCAAGGCTTTACAACTACTAAGATGTAAATACATTATTAAACTATACGATATGTTCCCGCGCGGCATGAGTCTTGTTCTCGTGCTAGAATATATGTACTCCGGATTATGGGAAATGTTGCACCAATGCGAGCATGAACTGACAATAGCTAGAGTCAAAACTTATGCAGAAATGTTATTAAAGGGAATGCGTTACATGCATGCTCATTACGTCATGCATAGGGATCTGAAACCAGCAAACCTGTTAATAAATCATGAGGGAATATTGAAAATAGCAGATCTAGGATTAGCGCGACTATACTGGCCTGAGGGTGGTAGGCCCTATTCACATCAAGTGGCCACAAGGTTAATACGCACTATATTATTTAATTAA
Protein
MDYDVNNYTVVGRIGEGAHGLVFKAKHNTTGREVALKKILIKNLEDGIPVHVMREIKALQLLRCKYIIKLYDMFPRGMSLVLVLEYMYSGLWEMLHQCEHELTIARVKTYAEMLLKGMRYMHAHYVMHRDLKPANLLINHEGILKIADLGLARLYWPEGGRPYSHQVATRLIRTILFN
Summary
Description
Involved in cell growth. Activates cdk2, a kinase involved in the control of the cell cycle, by phosphorylating residue 'Thr-160' (By similarity). Required for high-level Shh responses in the developing neural tube. Together with tbc1d32, controls the structure of the primary cilium by coordinating assembly of the ciliary membrane and axoneme, allowing gli2 to be properly activated in response to SHH signaling.
Involved in cell growth. Activates CDK2, a kinase involved in the control of the cell cycle, by phosphorylating residue 'Thr-160' (By similarity). Required for high-level Shh responses in the developing neural tube. Together with TBC1D32, controls the structure of the primary cilium by coordinating assembly of the ciliary membrane and axoneme, allowing GLI2 to be properly activated in response to SHH signaling.
Required for high-level Shh responses in the developing neural tube. Together with TBC1D32, controls the structure of the primary cilium by coordinating assembly of the ciliary membrane and axoneme, allowing GLI2 to be properly activated in response to SHH signaling. Involved in cell growth. Activates CDK2, a kinase involved in the control of the cell cycle, by phosphorylating residue 'Thr-160' (By similarity).
Involved in cell growth. Activates CDK2, a kinase involved in the control of the cell cycle, by phosphorylating residue 'Thr-160' (By similarity). Required for high-level Shh responses in the developing neural tube. Together with TBC1D32, controls the structure of the primary cilium by coordinating assembly of the ciliary membrane and axoneme, allowing GLI2 to be properly activated in response to SHH signaling.
Required for high-level Shh responses in the developing neural tube. Together with TBC1D32, controls the structure of the primary cilium by coordinating assembly of the ciliary membrane and axoneme, allowing GLI2 to be properly activated in response to SHH signaling. Involved in cell growth. Activates CDK2, a kinase involved in the control of the cell cycle, by phosphorylating residue 'Thr-160' (By similarity).
Catalytic Activity
ATP + L-seryl-[protein] = ADP + H(+) + O-phospho-L-seryl-[protein]
ATP + L-threonyl-[protein] = ADP + H(+) + O-phospho-L-threonyl-[protein]
ATP + L-threonyl-[protein] = ADP + H(+) + O-phospho-L-threonyl-[protein]
Subunit
Monomer. Interacts with tbc1d32.
Monomer (By similarity). Interacts with MAK (By similarity). Interacts with TBC1D32.
Monomer. Interacts with TBC1D32 and MAK.
Monomer (By similarity). Interacts with MAK (By similarity). Interacts with TBC1D32.
Monomer. Interacts with TBC1D32 and MAK.
Similarity
Belongs to the protein kinase superfamily.
Belongs to the protein kinase superfamily. CMGC Ser/Thr protein kinase family. CDC2/CDKX subfamily.
Belongs to the protein kinase superfamily. CMGC Ser/Thr protein kinase family. CDC2/CDKX subfamily.
Keywords
ATP-binding
Cell cycle
Cell division
Cell projection
Cilium
Complete proteome
Cytoplasm
Developmental protein
Kinase
Nucleotide-binding
Nucleus
Reference proteome
Transferase
Alternative splicing
Feature
chain Cyclin-dependent kinase 20
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
A0A2H1VQQ9
A0A2A4JFC8
A0A3S2LRP2
A0A194RLI1
A0A212F736
H9J605
+ More
A0A0L7KWI8 A0A2P8XZC4 A0A067QIA3 A0A2J7QLN6 E2C0C0 A0A0C9R231 K7IUI8 A0A232EX67 F4X6L5 A0A0C9PLU0 A0A195E7E4 A0A195F2F8 A0A158NLD3 A0A151XCJ8 A0A1B6BWK7 A0A1B6D7T3 A0A195CZN9 T1HNR4 E2ATA2 A0A026WJG0 A0A146MD59 A0A0A9WHZ5 A0A2Y9FL45 A0A1Y7VIW8 A0A341ACZ7 A0A2Y9MGK8 A0A340W849 A0A3Q3WV06 A0A3Q3CA41 A0A224XFJ1 W5LCM0 A8WIP6 A0A1W4WYD6 Q9JHU3-2 A0A3B4EIE8 A0A0V0JA06 A0A0X3PDD3 A0A340WDN7 A0A0P7WR33 A0A2D0R103 A0A3Q1J361 A0A3Q3K091 V8PAH0 A0A3B1JHK7 A8K6P8 A0A3Q0R3Q9 A0A3Q3LY45 G1KH48 A0A340WB01 A0A3B4EIG4 A0A3N0YRV9 A0A3Q3G5K9 A0A2K6F9B7 A0A2K6RYN7 A0A0L8FIL7 A0A2K5ECB0 W5NC09 A0A2G9SDB4 A0A2Y9FJ01 A0A1S3W275 A0A3Q4GZ80 A0A3B4F134 A0A3P8PQR0 A0A3P9DCQ6 Q9JHU3 A0A337S3C2 A0A341AF79 A0A3Q1JDT8 A0A341AF74 Q4KM34 A0A2K6F997 A0A2D0R1Q6 A0A2J8WNX6 A0A3Q3KB96 B4DFP8 A0A3B4WTA6 A0A3B4VC32 G3ICC3 A0A2K5KXN7 A0A091CP52 A0A3P9P7Q4 H0VCP1 A0A3P9ADU3 A0A2K5RK67 A0A1S3AT12 A0A3Q3M5W3 A0A1S3ASH7 A0A3B5PWL5 A0A0X3P0U7 D3K5N0 A0A2K6K5C0 A0A2K6NYZ6 A0A2Y9FJ58 A0A2Y9K4U7
A0A0L7KWI8 A0A2P8XZC4 A0A067QIA3 A0A2J7QLN6 E2C0C0 A0A0C9R231 K7IUI8 A0A232EX67 F4X6L5 A0A0C9PLU0 A0A195E7E4 A0A195F2F8 A0A158NLD3 A0A151XCJ8 A0A1B6BWK7 A0A1B6D7T3 A0A195CZN9 T1HNR4 E2ATA2 A0A026WJG0 A0A146MD59 A0A0A9WHZ5 A0A2Y9FL45 A0A1Y7VIW8 A0A341ACZ7 A0A2Y9MGK8 A0A340W849 A0A3Q3WV06 A0A3Q3CA41 A0A224XFJ1 W5LCM0 A8WIP6 A0A1W4WYD6 Q9JHU3-2 A0A3B4EIE8 A0A0V0JA06 A0A0X3PDD3 A0A340WDN7 A0A0P7WR33 A0A2D0R103 A0A3Q1J361 A0A3Q3K091 V8PAH0 A0A3B1JHK7 A8K6P8 A0A3Q0R3Q9 A0A3Q3LY45 G1KH48 A0A340WB01 A0A3B4EIG4 A0A3N0YRV9 A0A3Q3G5K9 A0A2K6F9B7 A0A2K6RYN7 A0A0L8FIL7 A0A2K5ECB0 W5NC09 A0A2G9SDB4 A0A2Y9FJ01 A0A1S3W275 A0A3Q4GZ80 A0A3B4F134 A0A3P8PQR0 A0A3P9DCQ6 Q9JHU3 A0A337S3C2 A0A341AF79 A0A3Q1JDT8 A0A341AF74 Q4KM34 A0A2K6F997 A0A2D0R1Q6 A0A2J8WNX6 A0A3Q3KB96 B4DFP8 A0A3B4WTA6 A0A3B4VC32 G3ICC3 A0A2K5KXN7 A0A091CP52 A0A3P9P7Q4 H0VCP1 A0A3P9ADU3 A0A2K5RK67 A0A1S3AT12 A0A3Q3M5W3 A0A1S3ASH7 A0A3B5PWL5 A0A0X3P0U7 D3K5N0 A0A2K6K5C0 A0A2K6NYZ6 A0A2Y9FJ58 A0A2Y9K4U7
EC Number
2.7.11.22
Pubmed
EMBL
ODYU01003861
SOQ43150.1
NWSH01001593
PCG70767.1
RSAL01000022
RVE52388.1
+ More
KQ460045 KPJ18175.1 AGBW02009936 OWR49543.1 BABH01029894 BABH01029895 JTDY01005048 KOB67431.1 PYGN01001128 PSN37361.1 KK853567 KDR06664.1 NEVH01013243 PNF29500.1 GL451770 EFN78579.1 GBYB01002060 JAG71827.1 AAZX01004049 NNAY01001765 OXU22976.1 GL888818 EGI57797.1 GBYB01002058 JAG71825.1 KQ979568 KYN20777.1 KQ981864 KYN34362.1 ADTU01019320 KQ982314 KYQ58018.1 GEDC01031632 GEDC01003575 JAS05666.1 JAS33723.1 GEDC01015566 GEDC01003859 JAS21732.1 JAS33439.1 KQ977115 KYN05604.1 ACPB03018039 GL442545 EFN63347.1 KK107185 QOIP01000012 EZA55816.1 RLU15907.1 GDHC01000955 JAQ17674.1 GBHO01039114 GBHO01039113 GBRD01015897 GBRD01015896 JAG04490.1 JAG04491.1 JAG49929.1 CT025616 GFTR01005191 JAW11235.1 BX571687 BC086697 AY005133 AY904369 BC031907 GEEE01021526 GEEE01012963 GEEE01001148 JAP62077.1 GEEE01013497 JAP49728.1 JARO02005892 KPP66074.1 AZIM01000454 ETE70966.1 AK291713 CH471089 BAF84402.1 EAW62747.1 RJVU01030786 ROL48408.1 KQ430924 KOF63485.1 AHAT01006438 AHAT01006439 KV924701 PIO38092.1 AANG04003916 BC098838 NDHI03003383 PNJ71459.1 AK294193 BAG57509.1 JH001908 RAZU01000141 EGW11400.1 RLQ69609.1 KN124574 KFO20774.1 AAKN02038453 GEEE01021643 GEEE01013999 JAP41582.1 GU373708 ADC38908.1
KQ460045 KPJ18175.1 AGBW02009936 OWR49543.1 BABH01029894 BABH01029895 JTDY01005048 KOB67431.1 PYGN01001128 PSN37361.1 KK853567 KDR06664.1 NEVH01013243 PNF29500.1 GL451770 EFN78579.1 GBYB01002060 JAG71827.1 AAZX01004049 NNAY01001765 OXU22976.1 GL888818 EGI57797.1 GBYB01002058 JAG71825.1 KQ979568 KYN20777.1 KQ981864 KYN34362.1 ADTU01019320 KQ982314 KYQ58018.1 GEDC01031632 GEDC01003575 JAS05666.1 JAS33723.1 GEDC01015566 GEDC01003859 JAS21732.1 JAS33439.1 KQ977115 KYN05604.1 ACPB03018039 GL442545 EFN63347.1 KK107185 QOIP01000012 EZA55816.1 RLU15907.1 GDHC01000955 JAQ17674.1 GBHO01039114 GBHO01039113 GBRD01015897 GBRD01015896 JAG04490.1 JAG04491.1 JAG49929.1 CT025616 GFTR01005191 JAW11235.1 BX571687 BC086697 AY005133 AY904369 BC031907 GEEE01021526 GEEE01012963 GEEE01001148 JAP62077.1 GEEE01013497 JAP49728.1 JARO02005892 KPP66074.1 AZIM01000454 ETE70966.1 AK291713 CH471089 BAF84402.1 EAW62747.1 RJVU01030786 ROL48408.1 KQ430924 KOF63485.1 AHAT01006438 AHAT01006439 KV924701 PIO38092.1 AANG04003916 BC098838 NDHI03003383 PNJ71459.1 AK294193 BAG57509.1 JH001908 RAZU01000141 EGW11400.1 RLQ69609.1 KN124574 KFO20774.1 AAKN02038453 GEEE01021643 GEEE01013999 JAP41582.1 GU373708 ADC38908.1
Proteomes
UP000218220
UP000283053
UP000053240
UP000007151
UP000005204
UP000037510
+ More
UP000245037 UP000027135 UP000235965 UP000008237 UP000002358 UP000215335 UP000007755 UP000078492 UP000078541 UP000005205 UP000075809 UP000078542 UP000015103 UP000000311 UP000053097 UP000279307 UP000248484 UP000000589 UP000252040 UP000248483 UP000265300 UP000261620 UP000264840 UP000018467 UP000000437 UP000192223 UP000261440 UP000034805 UP000221080 UP000265040 UP000261600 UP000261340 UP000261640 UP000001646 UP000261660 UP000233160 UP000233220 UP000053454 UP000233020 UP000018468 UP000079721 UP000261580 UP000261460 UP000265100 UP000265160 UP000011712 UP000002494 UP000261360 UP000261420 UP000001075 UP000273346 UP000233060 UP000028990 UP000242638 UP000005447 UP000265140 UP000233040 UP000002852 UP000233180 UP000233200 UP000248482
UP000245037 UP000027135 UP000235965 UP000008237 UP000002358 UP000215335 UP000007755 UP000078492 UP000078541 UP000005205 UP000075809 UP000078542 UP000015103 UP000000311 UP000053097 UP000279307 UP000248484 UP000000589 UP000252040 UP000248483 UP000265300 UP000261620 UP000264840 UP000018467 UP000000437 UP000192223 UP000261440 UP000034805 UP000221080 UP000265040 UP000261600 UP000261340 UP000261640 UP000001646 UP000261660 UP000233160 UP000233220 UP000053454 UP000233020 UP000018468 UP000079721 UP000261580 UP000261460 UP000265100 UP000265160 UP000011712 UP000002494 UP000261360 UP000261420 UP000001075 UP000273346 UP000233060 UP000028990 UP000242638 UP000005447 UP000265140 UP000233040 UP000002852 UP000233180 UP000233200 UP000248482
Interpro
SUPFAM
SSF56112
SSF56112
Gene 3D
CDD
ProteinModelPortal
A0A2H1VQQ9
A0A2A4JFC8
A0A3S2LRP2
A0A194RLI1
A0A212F736
H9J605
+ More
A0A0L7KWI8 A0A2P8XZC4 A0A067QIA3 A0A2J7QLN6 E2C0C0 A0A0C9R231 K7IUI8 A0A232EX67 F4X6L5 A0A0C9PLU0 A0A195E7E4 A0A195F2F8 A0A158NLD3 A0A151XCJ8 A0A1B6BWK7 A0A1B6D7T3 A0A195CZN9 T1HNR4 E2ATA2 A0A026WJG0 A0A146MD59 A0A0A9WHZ5 A0A2Y9FL45 A0A1Y7VIW8 A0A341ACZ7 A0A2Y9MGK8 A0A340W849 A0A3Q3WV06 A0A3Q3CA41 A0A224XFJ1 W5LCM0 A8WIP6 A0A1W4WYD6 Q9JHU3-2 A0A3B4EIE8 A0A0V0JA06 A0A0X3PDD3 A0A340WDN7 A0A0P7WR33 A0A2D0R103 A0A3Q1J361 A0A3Q3K091 V8PAH0 A0A3B1JHK7 A8K6P8 A0A3Q0R3Q9 A0A3Q3LY45 G1KH48 A0A340WB01 A0A3B4EIG4 A0A3N0YRV9 A0A3Q3G5K9 A0A2K6F9B7 A0A2K6RYN7 A0A0L8FIL7 A0A2K5ECB0 W5NC09 A0A2G9SDB4 A0A2Y9FJ01 A0A1S3W275 A0A3Q4GZ80 A0A3B4F134 A0A3P8PQR0 A0A3P9DCQ6 Q9JHU3 A0A337S3C2 A0A341AF79 A0A3Q1JDT8 A0A341AF74 Q4KM34 A0A2K6F997 A0A2D0R1Q6 A0A2J8WNX6 A0A3Q3KB96 B4DFP8 A0A3B4WTA6 A0A3B4VC32 G3ICC3 A0A2K5KXN7 A0A091CP52 A0A3P9P7Q4 H0VCP1 A0A3P9ADU3 A0A2K5RK67 A0A1S3AT12 A0A3Q3M5W3 A0A1S3ASH7 A0A3B5PWL5 A0A0X3P0U7 D3K5N0 A0A2K6K5C0 A0A2K6NYZ6 A0A2Y9FJ58 A0A2Y9K4U7
A0A0L7KWI8 A0A2P8XZC4 A0A067QIA3 A0A2J7QLN6 E2C0C0 A0A0C9R231 K7IUI8 A0A232EX67 F4X6L5 A0A0C9PLU0 A0A195E7E4 A0A195F2F8 A0A158NLD3 A0A151XCJ8 A0A1B6BWK7 A0A1B6D7T3 A0A195CZN9 T1HNR4 E2ATA2 A0A026WJG0 A0A146MD59 A0A0A9WHZ5 A0A2Y9FL45 A0A1Y7VIW8 A0A341ACZ7 A0A2Y9MGK8 A0A340W849 A0A3Q3WV06 A0A3Q3CA41 A0A224XFJ1 W5LCM0 A8WIP6 A0A1W4WYD6 Q9JHU3-2 A0A3B4EIE8 A0A0V0JA06 A0A0X3PDD3 A0A340WDN7 A0A0P7WR33 A0A2D0R103 A0A3Q1J361 A0A3Q3K091 V8PAH0 A0A3B1JHK7 A8K6P8 A0A3Q0R3Q9 A0A3Q3LY45 G1KH48 A0A340WB01 A0A3B4EIG4 A0A3N0YRV9 A0A3Q3G5K9 A0A2K6F9B7 A0A2K6RYN7 A0A0L8FIL7 A0A2K5ECB0 W5NC09 A0A2G9SDB4 A0A2Y9FJ01 A0A1S3W275 A0A3Q4GZ80 A0A3B4F134 A0A3P8PQR0 A0A3P9DCQ6 Q9JHU3 A0A337S3C2 A0A341AF79 A0A3Q1JDT8 A0A341AF74 Q4KM34 A0A2K6F997 A0A2D0R1Q6 A0A2J8WNX6 A0A3Q3KB96 B4DFP8 A0A3B4WTA6 A0A3B4VC32 G3ICC3 A0A2K5KXN7 A0A091CP52 A0A3P9P7Q4 H0VCP1 A0A3P9ADU3 A0A2K5RK67 A0A1S3AT12 A0A3Q3M5W3 A0A1S3ASH7 A0A3B5PWL5 A0A0X3P0U7 D3K5N0 A0A2K6K5C0 A0A2K6NYZ6 A0A2Y9FJ58 A0A2Y9K4U7
PDB
1V0O
E-value=5.07879e-29,
Score=314
Ontologies
KEGG
GO
PANTHER
Topology
Subcellular location
Nucleus
Cytoplasm
Cell projection
Cilium
Cytoplasm
Cell projection
Cilium
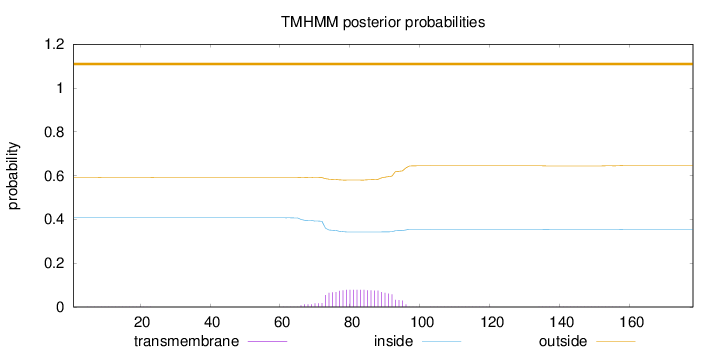
Length:
178
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.66476
Exp number, first 60 AAs:
0.00719999999999999
Total prob of N-in:
0.40752
outside
1 - 178
Population Genetic Test Statistics
Pi
289.722229
Theta
162.90995
Tajima's D
1.735876
CLR
0.125084
CSRT
0.83650817459127
Interpretation
Uncertain
