Gene
KWMTBOMO15198 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA004965
Annotation
uridine_diphosphate_glucosyltransferase_precursor_[Bombyx_mori]
Full name
UDP-glucuronosyltransferase
+ More
Ecdysteroid UDP-glucosyltransferase
Ecdysteroid UDP-glucosyltransferase
Location in the cell
PlasmaMembrane Reliability : 2.628
Sequence
CDS
ATGTGGTTAAAGTTTTTCTACTTCTTTAGTTTAGTTTTATGTCCGTGTTATGCGGCTCAAGGCGCTTCGATATTAGCAGTCTTCTCTTCACTTTCCTATTCAGATCATCTGGTTTTTCGGGGCTATGTGTCTCTCCTCGCTCAAAGAGGACATTCCGTTGTGGTCATGACTCCTTATCCAGGCGAGTTTCAATATCCGGAGGTAGAAAACATAATCGAGCTAGATGTTAGTCAAGAATCAGCACCGTTTTGGGAGGAGTACAAAAGGCTGATGACCAATACGGACGATTACTATCCGCGGCTTAAAGCACTGAACGAGCTCTCACTAAAAATTGCCATAGCGCAGCTGAAGTCCAAGCAAATGACGGCGCTGCTCATTAATCCTAATGTAAAATTCGACTTGGTAATAACTGAAGCAGATGTACCATTACTTTACGCTTTAGCTGACAAATACCAGACTCCACATATAACGATAAGTACATTTAATGGAAAAATTCACCAGTACGAAGCTAAAGGCAATCCCATTCACCCTATTTTGTATCCCGACGTGAACTCACTGAACTACGGTAATTTGACTCGTTGGCAGAAAATTATCGAATTCTACAGACACATACAAACGAAAACAGAGTTTTACAACAACTATTTGCCGTTATGCGATGTTGCCGCCAAAAAGATACTAGGATTGAAAAGAGATTTACAAGAAGTTGAATATGACATCGATATGTTGTTCATTGCCAGTAATCCGCTGTTAATCGGGAATAGACCGGTTGTACCTGCCATACAATTTGTGGATCGGATGCATATTAAACCAAGGATGAGTCTTCCGCAGAATTTACAATCATTGTTAGACTCTCAAACGAAGGGAGTAATTTACTTTAGCTTGGGAACTCTACAAGAGGCGGAGAAATTATCTGTTAAAACACTTCAAGTTTTCGCGGATGCTTTTAGAGAATTACCTTTCACAGTTTTGTGGAAGATTGGTAAAATGAGCACGTTAAAGCTATCTGATAACGTTATCACAGACGTATGGTTTCCGCAACAACAATTATTGGCACACAAAAACGTAAGGGCATTTATAACTCACGGCGGACCTAGATCTTTGGAAGAAGCACTATTTTATGAAGTACCAATCATAGGTTTTCCTCTTATCACATCTAGAAAGATATTTATAAGAGAGCTTACCAAGTACGGCGCTGGTGAAATCTTGGACCCCTTACACATTGACAAACAAACATTGAAACAGGTCATATCCACAGTGGCCACTGATGAAAAGTACAAAAAAGCGATAATTAAGCTTAAAGGTATGGTTGTCGATCCGTTGATTTCGGGTCCCGACAACGCTGTCTGGTGGACTGAATACGTTCTTCGTAACCGCGGCGCACAACATCTACGTTCACCAGTTGTTGGCGTTACCTTCATCAAATATTACATGTTGGACATTCTTACTTATATACTTGCAGTCGTTCTATTTCTGCTATATTTAACTTTTCTTGTGCTGAAATGTATTTACAGACGTTTACGAGCACGATTTGTGCTGAGAACTGGTCAAGGACCAGAAGGAAAATTCAAGGCTTTGTAA
Protein
MWLKFFYFFSLVLCPCYAAQGASILAVFSSLSYSDHLVFRGYVSLLAQRGHSVVVMTPYPGEFQYPEVENIIELDVSQESAPFWEEYKRLMTNTDDYYPRLKALNELSLKIAIAQLKSKQMTALLINPNVKFDLVITEADVPLLYALADKYQTPHITISTFNGKIHQYEAKGNPIHPILYPDVNSLNYGNLTRWQKIIEFYRHIQTKTEFYNNYLPLCDVAAKKILGLKRDLQEVEYDIDMLFIASNPLLIGNRPVVPAIQFVDRMHIKPRMSLPQNLQSLLDSQTKGVIYFSLGTLQEAEKLSVKTLQVFADAFRELPFTVLWKIGKMSTLKLSDNVITDVWFPQQQLLAHKNVRAFITHGGPRSLEEALFYEVPIIGFPLITSRKIFIRELTKYGAGEILDPLHIDKQTLKQVISTVATDEKYKKAIIKLKGMVVDPLISGPDNAVWWTEYVLRNRGAQHLRSPVVGVTFIKYYMLDILTYILAVVLFLLYLTFLVLKCIYRRLRARFVLRTGQGPEGKFKAL
Summary
Description
Catalyzes the transfer of glucose from UDP-glucose to ecdysteroids which are insect molting hormones.
Catalytic Activity
glucuronate acceptor + UDP-alpha-D-glucuronate = acceptor beta-D-glucuronoside + H(+) + UDP
Similarity
Belongs to the UDP-glycosyltransferase family.
Belongs to the Glu/Leu/Phe/Val dehydrogenases family.
Belongs to the Glu/Leu/Phe/Val dehydrogenases family.
Feature
chain UDP-glucuronosyltransferase
Uniprot
B5AU20
C0JKP8
A0A3G1T1A4
A0A2A4JG63
G9LPP8
A0A1E1WQJ5
+ More
A0A2H1VQM8 A0A0L7KWD8 A0A194RQ77 D2JLK9 A0A2H4WB71 A0A194PG03 A0A1L8D6G4 A0A212F721 A0A212EW38 A0A0N1I2R8 A0A1Y1M2S7 A0A0N0PE43 A0A2A4JXE4 V5GJ31 M4M1W9 A0A2W1BAD4 G9LPP6 A0A2H4WB54 A0A191T2J5 A0A1Y1M828 A0A1L8D6H9 A0A2H4WB68 A0A1Y1MKU1 A0A1W4XHT9 A0A024AGL3 A0A2I6RJL8 G9LPT6 A0A346TPQ3 A0A3S2NVG0 A0A191T1N0 A0A191T2J2 A0A2W1BL20 A0A139WAV2 G9LPP3 A0A191T2G3 A0A2H1WTF5 A0A024AG00 A0A2R4FXG6 A0A2H1VUS4 A0A2H4WB70 A0A2H4WB73 A0A2W1BQP4 D2A215 A0A191T1L3 G9LPP7 A0A2R4FXJ6 H9JWC0 A0A2H1VUM4 A0A1W4XIA4 A0A191T1K8 I4DM75 A0A024AH14 A0A0N1IMZ0 A0A2W1B3V4 A0A2W1BEA5 G9LPP5 A0A1Y1LVX9 B7TZA3 G9LPS9 A0A2H1VU94 A0A2A4JWQ0 A0A191T1K4 G9LPS5 A0A2H4WB72 A0A1Y1MFU9 A0A2H1WSA7 A0A1Y1KEF7 G9LPN5 A0A2W1BA70 A0A140EA35 A0A191T1L2 Q1HPX7 A0A1L8D6H2 G9LPS6 A0A0N0P9C8 A0A1L8D6T8 V5GKI9 G9LPN8 A0A2W1BE55 A0A2W1B475 A0A2W1B9W6 G9LPT1 G9LPN6 A0A1L8D6G1 A0A191T1L8 A0A1Y1LUT9 G9LPN9 A0A2H4WB83 A0A191T1M4 A0A1Y1NHT4 A0A2W1BBH1 A0A2W1B8G2 A0A2W1B8D3
A0A2H1VQM8 A0A0L7KWD8 A0A194RQ77 D2JLK9 A0A2H4WB71 A0A194PG03 A0A1L8D6G4 A0A212F721 A0A212EW38 A0A0N1I2R8 A0A1Y1M2S7 A0A0N0PE43 A0A2A4JXE4 V5GJ31 M4M1W9 A0A2W1BAD4 G9LPP6 A0A2H4WB54 A0A191T2J5 A0A1Y1M828 A0A1L8D6H9 A0A2H4WB68 A0A1Y1MKU1 A0A1W4XHT9 A0A024AGL3 A0A2I6RJL8 G9LPT6 A0A346TPQ3 A0A3S2NVG0 A0A191T1N0 A0A191T2J2 A0A2W1BL20 A0A139WAV2 G9LPP3 A0A191T2G3 A0A2H1WTF5 A0A024AG00 A0A2R4FXG6 A0A2H1VUS4 A0A2H4WB70 A0A2H4WB73 A0A2W1BQP4 D2A215 A0A191T1L3 G9LPP7 A0A2R4FXJ6 H9JWC0 A0A2H1VUM4 A0A1W4XIA4 A0A191T1K8 I4DM75 A0A024AH14 A0A0N1IMZ0 A0A2W1B3V4 A0A2W1BEA5 G9LPP5 A0A1Y1LVX9 B7TZA3 G9LPS9 A0A2H1VU94 A0A2A4JWQ0 A0A191T1K4 G9LPS5 A0A2H4WB72 A0A1Y1MFU9 A0A2H1WSA7 A0A1Y1KEF7 G9LPN5 A0A2W1BA70 A0A140EA35 A0A191T1L2 Q1HPX7 A0A1L8D6H2 G9LPS6 A0A0N0P9C8 A0A1L8D6T8 V5GKI9 G9LPN8 A0A2W1BE55 A0A2W1B475 A0A2W1B9W6 G9LPT1 G9LPN6 A0A1L8D6G1 A0A191T1L8 A0A1Y1LUT9 G9LPN9 A0A2H4WB83 A0A191T1M4 A0A1Y1NHT4 A0A2W1BBH1 A0A2W1B8G2 A0A2W1B8D3
EC Number
2.4.1.17
2.4.1.-
2.4.1.-
Pubmed
EMBL
BABH01029895
EU873318
JQ070244
ACF57674.1
AEW43160.1
FJ237534
+ More
ACN39598.1 MG846900 AXY94752.1 NWSH01001593 PCG70766.1 JQ070205 AEW43121.1 GDQN01001867 JAT89187.1 ODYU01003861 SOQ43151.1 JTDY01005048 KOB67430.1 KQ460045 KPJ18176.1 GQ915325 ACZ97419.2 KY202940 AUC64284.1 KQ459604 KPI92316.1 GEYN01000080 JAV02049.1 AGBW02009936 OWR49542.1 AGBW02012096 OWR45677.1 KQ459547 KPJ00048.1 GEZM01042572 JAV79781.1 KQ459937 KPJ19195.1 NWSH01000435 PCG76446.1 GALX01004392 JAB64074.1 JQ747500 AGG36457.1 KZ150193 PZC72372.1 JQ070203 AEW43119.1 KY202924 AUC64268.1 KU680289 ANI21995.1 GEZM01037675 JAV81989.1 GEYN01000078 JAV02051.1 KY202939 AUC64283.1 GEZM01030332 GEZM01030328 JAV85628.1 KF777112 AHY99683.1 KY304474 AUN86405.1 JQ070243 AEW43159.1 MH124167 AXU41563.1 RSAL01000157 RVE45575.1 KU680291 ANI21997.1 KU680284 ANI21990.1 PZC72373.1 KQ971380 KYB25037.1 JQ070200 AEW43116.1 KU680290 ANI21996.1 ODYU01010933 SOQ56350.1 KF777109 AHY99680.1 MF034854 AVT42220.1 ODYU01004546 SOQ44548.1 KY202938 AUC64282.1 KY202933 AUC64277.1 KZ149974 PZC75964.1 KQ971338 EFA01500.1 KU680288 ANI21994.1 JQ070204 AEW43120.1 MF034855 AVT42221.1 BABH01029259 SOQ44549.1 KU680282 ANI21988.1 AK402393 BAM19015.1 KF777111 AHY99682.1 KPJ00054.1 KZ150360 PZC71192.1 PZC71190.1 JQ070202 AEW43118.1 GEZM01049908 JAV75766.1 FJ418685 ACJ48963.1 BABH01029271 JQ070236 AEW43152.1 ODYU01004485 SOQ44409.1 PCG76447.1 KU680283 ANI21989.1 JQ070232 AEW43148.1 KY202941 AUC64285.1 GEZM01032416 JAV84594.1 ODYU01010294 SOQ55304.1 GEZM01086036 JAV59829.1 JQ070192 AEW43108.1 PZC71195.1 KU214506 AMK97472.1 KU680287 ANI21993.1 DQ443275 ABF51364.1 GEYN01000077 JAV02052.1 JQ070233 AEW43149.1 KPJ00042.1 GEYN01000079 JAV02050.1 GALX01003882 JAB64584.1 JQ070195 AEW43111.1 PZC71196.1 PZC71189.1 PZC72369.1 JQ070238 AEW43154.1 JQ070193 AEW43109.1 GEYN01000091 JAV02038.1 KU680281 ANI21987.1 GEZM01049909 JAV75765.1 JQ070196 AEW43112.1 KY202943 AUC64287.1 KU680286 ANI21992.1 GEZM01006626 JAV95676.1 PZC71194.1 PZC71201.1 PZC71191.1
ACN39598.1 MG846900 AXY94752.1 NWSH01001593 PCG70766.1 JQ070205 AEW43121.1 GDQN01001867 JAT89187.1 ODYU01003861 SOQ43151.1 JTDY01005048 KOB67430.1 KQ460045 KPJ18176.1 GQ915325 ACZ97419.2 KY202940 AUC64284.1 KQ459604 KPI92316.1 GEYN01000080 JAV02049.1 AGBW02009936 OWR49542.1 AGBW02012096 OWR45677.1 KQ459547 KPJ00048.1 GEZM01042572 JAV79781.1 KQ459937 KPJ19195.1 NWSH01000435 PCG76446.1 GALX01004392 JAB64074.1 JQ747500 AGG36457.1 KZ150193 PZC72372.1 JQ070203 AEW43119.1 KY202924 AUC64268.1 KU680289 ANI21995.1 GEZM01037675 JAV81989.1 GEYN01000078 JAV02051.1 KY202939 AUC64283.1 GEZM01030332 GEZM01030328 JAV85628.1 KF777112 AHY99683.1 KY304474 AUN86405.1 JQ070243 AEW43159.1 MH124167 AXU41563.1 RSAL01000157 RVE45575.1 KU680291 ANI21997.1 KU680284 ANI21990.1 PZC72373.1 KQ971380 KYB25037.1 JQ070200 AEW43116.1 KU680290 ANI21996.1 ODYU01010933 SOQ56350.1 KF777109 AHY99680.1 MF034854 AVT42220.1 ODYU01004546 SOQ44548.1 KY202938 AUC64282.1 KY202933 AUC64277.1 KZ149974 PZC75964.1 KQ971338 EFA01500.1 KU680288 ANI21994.1 JQ070204 AEW43120.1 MF034855 AVT42221.1 BABH01029259 SOQ44549.1 KU680282 ANI21988.1 AK402393 BAM19015.1 KF777111 AHY99682.1 KPJ00054.1 KZ150360 PZC71192.1 PZC71190.1 JQ070202 AEW43118.1 GEZM01049908 JAV75766.1 FJ418685 ACJ48963.1 BABH01029271 JQ070236 AEW43152.1 ODYU01004485 SOQ44409.1 PCG76447.1 KU680283 ANI21989.1 JQ070232 AEW43148.1 KY202941 AUC64285.1 GEZM01032416 JAV84594.1 ODYU01010294 SOQ55304.1 GEZM01086036 JAV59829.1 JQ070192 AEW43108.1 PZC71195.1 KU214506 AMK97472.1 KU680287 ANI21993.1 DQ443275 ABF51364.1 GEYN01000077 JAV02052.1 JQ070233 AEW43149.1 KPJ00042.1 GEYN01000079 JAV02050.1 GALX01003882 JAB64584.1 JQ070195 AEW43111.1 PZC71196.1 PZC71189.1 PZC72369.1 JQ070238 AEW43154.1 JQ070193 AEW43109.1 GEYN01000091 JAV02038.1 KU680281 ANI21987.1 GEZM01049909 JAV75765.1 JQ070196 AEW43112.1 KY202943 AUC64287.1 KU680286 ANI21992.1 GEZM01006626 JAV95676.1 PZC71194.1 PZC71201.1 PZC71191.1
Proteomes
Interpro
IPR002213
UDP_glucos_trans
+ More
IPR035595 UDP_glycos_trans_CS
IPR008485 JAMP
IPR006326 UDPGT_MGT
IPR016224 Ecdysteroid_UDP-Glc_Trfase
IPR036291 NAD(P)-bd_dom_sf
IPR033524 Glu/Leu/Phe/Val_DH_AS
IPR006096 Glu/Leu/Phe/Val_DH_C
IPR006095 Glu/Leu/Phe/Val_DH
IPR006097 Glu/Leu/Phe/Val_DH_dimer_dom
IPR033922 NAD_bind_Glu_DH
IPR035595 UDP_glycos_trans_CS
IPR008485 JAMP
IPR006326 UDPGT_MGT
IPR016224 Ecdysteroid_UDP-Glc_Trfase
IPR036291 NAD(P)-bd_dom_sf
IPR033524 Glu/Leu/Phe/Val_DH_AS
IPR006096 Glu/Leu/Phe/Val_DH_C
IPR006095 Glu/Leu/Phe/Val_DH
IPR006097 Glu/Leu/Phe/Val_DH_dimer_dom
IPR033922 NAD_bind_Glu_DH
SUPFAM
SSF51735
SSF51735
ProteinModelPortal
B5AU20
C0JKP8
A0A3G1T1A4
A0A2A4JG63
G9LPP8
A0A1E1WQJ5
+ More
A0A2H1VQM8 A0A0L7KWD8 A0A194RQ77 D2JLK9 A0A2H4WB71 A0A194PG03 A0A1L8D6G4 A0A212F721 A0A212EW38 A0A0N1I2R8 A0A1Y1M2S7 A0A0N0PE43 A0A2A4JXE4 V5GJ31 M4M1W9 A0A2W1BAD4 G9LPP6 A0A2H4WB54 A0A191T2J5 A0A1Y1M828 A0A1L8D6H9 A0A2H4WB68 A0A1Y1MKU1 A0A1W4XHT9 A0A024AGL3 A0A2I6RJL8 G9LPT6 A0A346TPQ3 A0A3S2NVG0 A0A191T1N0 A0A191T2J2 A0A2W1BL20 A0A139WAV2 G9LPP3 A0A191T2G3 A0A2H1WTF5 A0A024AG00 A0A2R4FXG6 A0A2H1VUS4 A0A2H4WB70 A0A2H4WB73 A0A2W1BQP4 D2A215 A0A191T1L3 G9LPP7 A0A2R4FXJ6 H9JWC0 A0A2H1VUM4 A0A1W4XIA4 A0A191T1K8 I4DM75 A0A024AH14 A0A0N1IMZ0 A0A2W1B3V4 A0A2W1BEA5 G9LPP5 A0A1Y1LVX9 B7TZA3 G9LPS9 A0A2H1VU94 A0A2A4JWQ0 A0A191T1K4 G9LPS5 A0A2H4WB72 A0A1Y1MFU9 A0A2H1WSA7 A0A1Y1KEF7 G9LPN5 A0A2W1BA70 A0A140EA35 A0A191T1L2 Q1HPX7 A0A1L8D6H2 G9LPS6 A0A0N0P9C8 A0A1L8D6T8 V5GKI9 G9LPN8 A0A2W1BE55 A0A2W1B475 A0A2W1B9W6 G9LPT1 G9LPN6 A0A1L8D6G1 A0A191T1L8 A0A1Y1LUT9 G9LPN9 A0A2H4WB83 A0A191T1M4 A0A1Y1NHT4 A0A2W1BBH1 A0A2W1B8G2 A0A2W1B8D3
A0A2H1VQM8 A0A0L7KWD8 A0A194RQ77 D2JLK9 A0A2H4WB71 A0A194PG03 A0A1L8D6G4 A0A212F721 A0A212EW38 A0A0N1I2R8 A0A1Y1M2S7 A0A0N0PE43 A0A2A4JXE4 V5GJ31 M4M1W9 A0A2W1BAD4 G9LPP6 A0A2H4WB54 A0A191T2J5 A0A1Y1M828 A0A1L8D6H9 A0A2H4WB68 A0A1Y1MKU1 A0A1W4XHT9 A0A024AGL3 A0A2I6RJL8 G9LPT6 A0A346TPQ3 A0A3S2NVG0 A0A191T1N0 A0A191T2J2 A0A2W1BL20 A0A139WAV2 G9LPP3 A0A191T2G3 A0A2H1WTF5 A0A024AG00 A0A2R4FXG6 A0A2H1VUS4 A0A2H4WB70 A0A2H4WB73 A0A2W1BQP4 D2A215 A0A191T1L3 G9LPP7 A0A2R4FXJ6 H9JWC0 A0A2H1VUM4 A0A1W4XIA4 A0A191T1K8 I4DM75 A0A024AH14 A0A0N1IMZ0 A0A2W1B3V4 A0A2W1BEA5 G9LPP5 A0A1Y1LVX9 B7TZA3 G9LPS9 A0A2H1VU94 A0A2A4JWQ0 A0A191T1K4 G9LPS5 A0A2H4WB72 A0A1Y1MFU9 A0A2H1WSA7 A0A1Y1KEF7 G9LPN5 A0A2W1BA70 A0A140EA35 A0A191T1L2 Q1HPX7 A0A1L8D6H2 G9LPS6 A0A0N0P9C8 A0A1L8D6T8 V5GKI9 G9LPN8 A0A2W1BE55 A0A2W1B475 A0A2W1B9W6 G9LPT1 G9LPN6 A0A1L8D6G1 A0A191T1L8 A0A1Y1LUT9 G9LPN9 A0A2H4WB83 A0A191T1M4 A0A1Y1NHT4 A0A2W1BBH1 A0A2W1B8G2 A0A2W1B8D3
PDB
2O6L
E-value=7.54592e-17,
Score=215
Ontologies
PATHWAY
00040
Pentose and glucuronate interconversions - Bombyx mori (domestic silkworm)
00053 Ascorbate and aldarate metabolism - Bombyx mori (domestic silkworm)
00830 Retinol metabolism - Bombyx mori (domestic silkworm)
00860 Porphyrin and chlorophyll metabolism - Bombyx mori (domestic silkworm)
00980 Metabolism of xenobiotics by cytochrome P450 - Bombyx mori (domestic silkworm)
00982 Drug metabolism - cytochrome P450 - Bombyx mori (domestic silkworm)
00983 Drug metabolism - other enzymes - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
00053 Ascorbate and aldarate metabolism - Bombyx mori (domestic silkworm)
00830 Retinol metabolism - Bombyx mori (domestic silkworm)
00860 Porphyrin and chlorophyll metabolism - Bombyx mori (domestic silkworm)
00980 Metabolism of xenobiotics by cytochrome P450 - Bombyx mori (domestic silkworm)
00982 Drug metabolism - cytochrome P450 - Bombyx mori (domestic silkworm)
00983 Drug metabolism - other enzymes - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
GO
PANTHER
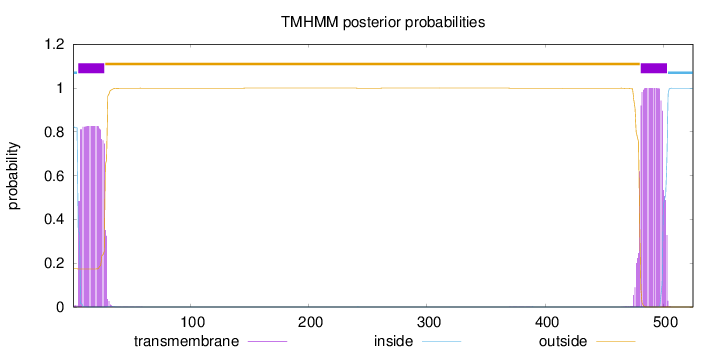
Topology
Subcellular location
Membrane
SignalP
Position: 1 - 18,
Likelihood: 0.932664

Length:
525
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
40.91694
Exp number, first 60 AAs:
18.84436
Total prob of N-in:
0.82575
POSSIBLE N-term signal
sequence
inside
1 - 4
TMhelix
5 - 27
outside
28 - 480
TMhelix
481 - 503
inside
504 - 525
Population Genetic Test Statistics
Pi
218.657884
Theta
177.48336
Tajima's D
0.699852
CLR
0.157634
CSRT
0.574971251437428
Interpretation
Uncertain
