Gene
KWMTBOMO15197
Pre Gene Modal
BGIBMGA004966
Annotation
PREDICTED:_JNK1/MAPK8-associated_membrane_protein_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 4.987
Sequence
CDS
ATGGCAGCTGTAAGATCGTGTCCAGGACTGTATTGTGGTCGTATTGAACTCGAAGATGGGTCGTGGAGTGACTGTGGGGCGTGTCCGCGAGGTTTCAGGACAAACGCATCAAGTTACTGCATGCCGTGCGTTGATGAACCCTCTCTATATGACTGGCAGTACCTTGGCTTTATGGTCCTCCTGCCCATGGTTTTGCATTGGTTTTTCATTGACATGGTCACCAGGGGTAAAGCCGAGAATGGCGTTATCATACAACACATGTGTGCCTTTTTGGAGGTCATATCAGGAACACTTGCCGCATTGTTAGTGCTGCCTCCGCCTGGGTCAGTCTCACTATATATGTGTTCACCAAAAGCACTTTCGGATTGGTATACACTGTTGCATAATCCACAGCCAGATTATAAAGAGATGTTGCATTGTACTCAGGAAGCAGTGTATCCTTTATATACAATAGTTTTTCTGATTTATGCATTCAGCTTATTAATGACCATAACATTAAGACCATGGTTACTGGCCTGGAATGCACCCAATTCTGGTAAAAAGGCAATATATTGTGCTTTATATTTTCATCCTATACTTGTATTGATTCATACTGTTGCTGCCGGTTTAATTTATTGCTCATTCCCCTACATAGTAATAATAATATCAATGATGACATCAGCATCGCATTTCTCAATTAAATCAGAACAGTCTGCGCCAGCACTATTGATTTCTTCTATCACTAATACACGGAATTTAATTATTCTCTTTGGACACTGGTTGGTCCATGCATATGGCATATTATCACTAACGGGGTTTAAAGAATTATGGTATTTGTCGTTAGTGCCCGCTCCGGCTATGTTTTATATTGTGACGGCACAATTCACAGATCCCTTGAAGATACATGGTGATTGA
Protein
MAAVRSCPGLYCGRIELEDGSWSDCGACPRGFRTNASSYCMPCVDEPSLYDWQYLGFMVLLPMVLHWFFIDMVTRGKAENGVIIQHMCAFLEVISGTLAALLVLPPPGSVSLYMCSPKALSDWYTLLHNPQPDYKEMLHCTQEAVYPLYTIVFLIYAFSLLMTITLRPWLLAWNAPNSGKKAIYCALYFHPILVLIHTVAAGLIYCSFPYIVIIISMMTSASHFSIKSEQSAPALLISSITNTRNLIILFGHWLVHAYGILSLTGFKELWYLSLVPAPAMFYIVTAQFTDPLKIHGD
Summary
Uniprot
H9J625
A0A3S2M6Y1
A0A2A4JF96
A0A194RKV8
A0A194PMH3
A0A212F727
+ More
A0A2H1VQM8 A0A212EW10 A0A0A9XNA6 A0A026WZ80 A0A2A3EPZ4 V9IJA0 A0A2R7WCA8 A0A067RTL4 A0A195F1V2 A0A232FDI7 A0A151XBJ3 E9IF96 A0A0V0G8P1 R4G436 E2B6I9 A0A151JT30 F4WFW0 K7J316 A0A158NL56 E2AI00 A0A195BH09 A0A0L7QTM9 R4WSH1 A0A023FA46 A0A195C023 A0A0P4VIQ4 A0A2P8YKD3 A0A1B6CXA4 T1J971 A0A1W4X7I5 A0A088AB43 A0A1B6KTR5 A0A1W4X7C1 A0A1B6GVV2 A0A1B0CJ92 A0A1Y1MNA9 A0A154P3D7 A0A1L8D928 A0A0M8ZVT1 A0A0K8TM03 D6WT95 U5EYB0 V5HWY2 A0A2P6KEU0 A0A1Z5LEE2 A0A2R5L753 A0A182HA92 A0A023ENZ0 A0A0P4WNI5 A0A182G0Y9 A0A2L2YF39 Q17EV3 A0A1Q3F6L7 A0A210PXI3 B0WYU5 A0A1S3J776 A0A1S3J6X0 A0A1W4X7D6 W5MSE2 A0A182JE75 A0A0K2TXC8 A0A3B5K4H1 D3PIB2 A0A131XCF8 A0A3B4FNW5 I3JZN4 A0A3P8NVW3 A0A3P9CKS1 A0A182JRL3 L7M342 A0A3Q1B8Z3 A0A3P8SEQ6 F1R9P8 Q6DHM8 A0A182PH23 A0A3Q1EKZ0 A0A224Z195 H3CS83 A0A182TP44 C3XZ44 A0A1C7CZT6 A0A2C9GQT7 A0A182ULQ9 Q7PXL1 Q4SMG2 A0A131YPY2 H3BHC8 A0A3N0XVL8 A0A182LTL3 H3BHC9 I3JZN5 S4RFW1 A0A182RF22 S4RFW0
A0A2H1VQM8 A0A212EW10 A0A0A9XNA6 A0A026WZ80 A0A2A3EPZ4 V9IJA0 A0A2R7WCA8 A0A067RTL4 A0A195F1V2 A0A232FDI7 A0A151XBJ3 E9IF96 A0A0V0G8P1 R4G436 E2B6I9 A0A151JT30 F4WFW0 K7J316 A0A158NL56 E2AI00 A0A195BH09 A0A0L7QTM9 R4WSH1 A0A023FA46 A0A195C023 A0A0P4VIQ4 A0A2P8YKD3 A0A1B6CXA4 T1J971 A0A1W4X7I5 A0A088AB43 A0A1B6KTR5 A0A1W4X7C1 A0A1B6GVV2 A0A1B0CJ92 A0A1Y1MNA9 A0A154P3D7 A0A1L8D928 A0A0M8ZVT1 A0A0K8TM03 D6WT95 U5EYB0 V5HWY2 A0A2P6KEU0 A0A1Z5LEE2 A0A2R5L753 A0A182HA92 A0A023ENZ0 A0A0P4WNI5 A0A182G0Y9 A0A2L2YF39 Q17EV3 A0A1Q3F6L7 A0A210PXI3 B0WYU5 A0A1S3J776 A0A1S3J6X0 A0A1W4X7D6 W5MSE2 A0A182JE75 A0A0K2TXC8 A0A3B5K4H1 D3PIB2 A0A131XCF8 A0A3B4FNW5 I3JZN4 A0A3P8NVW3 A0A3P9CKS1 A0A182JRL3 L7M342 A0A3Q1B8Z3 A0A3P8SEQ6 F1R9P8 Q6DHM8 A0A182PH23 A0A3Q1EKZ0 A0A224Z195 H3CS83 A0A182TP44 C3XZ44 A0A1C7CZT6 A0A2C9GQT7 A0A182ULQ9 Q7PXL1 Q4SMG2 A0A131YPY2 H3BHC8 A0A3N0XVL8 A0A182LTL3 H3BHC9 I3JZN5 S4RFW1 A0A182RF22 S4RFW0
Pubmed
19121390
26354079
22118469
25401762
24508170
30249741
+ More
24845553 28648823 21282665 20798317 21719571 20075255 21347285 23691247 25474469 27129103 29403074 28004739 26369729 18362917 19820115 25765539 28528879 26483478 24945155 26561354 17510324 28812685 21551351 28049606 25186727 25576852 23594743 28797301 15496914 18563158 20966253 12364791 14747013 17210077 26830274 9215903
24845553 28648823 21282665 20798317 21719571 20075255 21347285 23691247 25474469 27129103 29403074 28004739 26369729 18362917 19820115 25765539 28528879 26483478 24945155 26561354 17510324 28812685 21551351 28049606 25186727 25576852 23594743 28797301 15496914 18563158 20966253 12364791 14747013 17210077 26830274 9215903
EMBL
BABH01029895
RSAL01000022
RVE52386.1
NWSH01001593
PCG70765.1
KQ460045
+ More
KPJ18177.1 KQ459604 KPI92315.1 AGBW02009936 OWR49541.1 ODYU01003861 SOQ43151.1 AGBW02012096 OWR45676.1 GBHO01021402 GBHO01021401 GBRD01018096 JAG22202.1 JAG22203.1 JAG47731.1 KK107078 QOIP01000011 EZA60454.1 RLU16630.1 KZ288194 PBC33835.1 JR049914 AEY61120.1 KK854446 PTY15925.1 KK852425 KDR24140.1 KQ981866 KYN34074.1 NNAY01000371 OXU28831.1 KQ982320 KYQ57724.1 GL762798 EFZ20754.1 GECL01001676 JAP04448.1 ACPB03008832 GAHY01001131 JAA76379.1 GL445975 EFN88681.1 KQ978530 KYN30311.1 GL888128 EGI66727.1 ADTU01019257 GL439621 EFN66908.1 KQ976488 KYM83513.1 KQ414741 KOC61980.1 AK417652 BAN20867.1 GBBI01000914 JAC17798.1 KQ978501 KYM93521.1 GDKW01001837 JAI54758.1 PYGN01000531 PSN44720.1 GEDC01022225 GEDC01019455 GEDC01019232 GEDC01010358 JAS15073.1 JAS17843.1 JAS18066.1 JAS26940.1 JH431970 GEBQ01025135 JAT14842.1 GECZ01003306 JAS66463.1 AJWK01014318 GEZM01026232 JAV87129.1 KQ434809 KZC06343.1 GFDF01011121 JAV02963.1 KQ435848 KOX71036.1 GDAI01002430 JAI15173.1 KQ971352 EFA06682.1 GANO01002113 JAB57758.1 GANP01005485 JAB78983.1 MWRG01013954 PRD24840.1 GFJQ02001245 JAW05725.1 GGLE01001210 MBY05336.1 JXUM01031157 KQ560911 KXJ80376.1 GAPW01002922 JAC10676.1 GDRN01057610 JAI65669.1 JXUM01037699 KQ561138 KXJ79598.1 IAAA01016968 LAA05835.1 CH477279 EAT44998.1 GFDL01011834 JAV23211.1 NEDP02005416 OWF41193.1 DS232199 EDS37205.1 AHAT01003879 HACA01013134 CDW30495.1 BT121368 ADD38298.1 GEFH01004519 JAP64062.1 AERX01049695 AERX01049696 AERX01049697 GACK01007525 JAA57509.1 BX004994 BC075939 AAH75939.1 GFPF01009217 MAA20363.1 GG666475 EEN66608.1 APCN01000875 AAAB01008987 EAA00861.3 CAAE01014551 CAF98170.1 GEDV01008401 JAP80156.1 AFYH01002560 AFYH01002561 RJVU01059333 ROJ78770.1 AXCM01000128
KPJ18177.1 KQ459604 KPI92315.1 AGBW02009936 OWR49541.1 ODYU01003861 SOQ43151.1 AGBW02012096 OWR45676.1 GBHO01021402 GBHO01021401 GBRD01018096 JAG22202.1 JAG22203.1 JAG47731.1 KK107078 QOIP01000011 EZA60454.1 RLU16630.1 KZ288194 PBC33835.1 JR049914 AEY61120.1 KK854446 PTY15925.1 KK852425 KDR24140.1 KQ981866 KYN34074.1 NNAY01000371 OXU28831.1 KQ982320 KYQ57724.1 GL762798 EFZ20754.1 GECL01001676 JAP04448.1 ACPB03008832 GAHY01001131 JAA76379.1 GL445975 EFN88681.1 KQ978530 KYN30311.1 GL888128 EGI66727.1 ADTU01019257 GL439621 EFN66908.1 KQ976488 KYM83513.1 KQ414741 KOC61980.1 AK417652 BAN20867.1 GBBI01000914 JAC17798.1 KQ978501 KYM93521.1 GDKW01001837 JAI54758.1 PYGN01000531 PSN44720.1 GEDC01022225 GEDC01019455 GEDC01019232 GEDC01010358 JAS15073.1 JAS17843.1 JAS18066.1 JAS26940.1 JH431970 GEBQ01025135 JAT14842.1 GECZ01003306 JAS66463.1 AJWK01014318 GEZM01026232 JAV87129.1 KQ434809 KZC06343.1 GFDF01011121 JAV02963.1 KQ435848 KOX71036.1 GDAI01002430 JAI15173.1 KQ971352 EFA06682.1 GANO01002113 JAB57758.1 GANP01005485 JAB78983.1 MWRG01013954 PRD24840.1 GFJQ02001245 JAW05725.1 GGLE01001210 MBY05336.1 JXUM01031157 KQ560911 KXJ80376.1 GAPW01002922 JAC10676.1 GDRN01057610 JAI65669.1 JXUM01037699 KQ561138 KXJ79598.1 IAAA01016968 LAA05835.1 CH477279 EAT44998.1 GFDL01011834 JAV23211.1 NEDP02005416 OWF41193.1 DS232199 EDS37205.1 AHAT01003879 HACA01013134 CDW30495.1 BT121368 ADD38298.1 GEFH01004519 JAP64062.1 AERX01049695 AERX01049696 AERX01049697 GACK01007525 JAA57509.1 BX004994 BC075939 AAH75939.1 GFPF01009217 MAA20363.1 GG666475 EEN66608.1 APCN01000875 AAAB01008987 EAA00861.3 CAAE01014551 CAF98170.1 GEDV01008401 JAP80156.1 AFYH01002560 AFYH01002561 RJVU01059333 ROJ78770.1 AXCM01000128
Proteomes
UP000005204
UP000283053
UP000218220
UP000053240
UP000053268
UP000007151
+ More
UP000053097 UP000279307 UP000242457 UP000027135 UP000078541 UP000215335 UP000075809 UP000015103 UP000008237 UP000078492 UP000007755 UP000002358 UP000005205 UP000000311 UP000078540 UP000053825 UP000078542 UP000245037 UP000192223 UP000005203 UP000092461 UP000076502 UP000053105 UP000007266 UP000069940 UP000249989 UP000008820 UP000242188 UP000002320 UP000085678 UP000018468 UP000075880 UP000005226 UP000261460 UP000005207 UP000265100 UP000265160 UP000075881 UP000257160 UP000265080 UP000000437 UP000075885 UP000257200 UP000007303 UP000075902 UP000001554 UP000075882 UP000075840 UP000075903 UP000007062 UP000008672 UP000075883 UP000245300 UP000075900
UP000053097 UP000279307 UP000242457 UP000027135 UP000078541 UP000215335 UP000075809 UP000015103 UP000008237 UP000078492 UP000007755 UP000002358 UP000005205 UP000000311 UP000078540 UP000053825 UP000078542 UP000245037 UP000192223 UP000005203 UP000092461 UP000076502 UP000053105 UP000007266 UP000069940 UP000249989 UP000008820 UP000242188 UP000002320 UP000085678 UP000018468 UP000075880 UP000005226 UP000261460 UP000005207 UP000265100 UP000265160 UP000075881 UP000257160 UP000265080 UP000000437 UP000075885 UP000257200 UP000007303 UP000075902 UP000001554 UP000075882 UP000075840 UP000075903 UP000007062 UP000008672 UP000075883 UP000245300 UP000075900
ProteinModelPortal
H9J625
A0A3S2M6Y1
A0A2A4JF96
A0A194RKV8
A0A194PMH3
A0A212F727
+ More
A0A2H1VQM8 A0A212EW10 A0A0A9XNA6 A0A026WZ80 A0A2A3EPZ4 V9IJA0 A0A2R7WCA8 A0A067RTL4 A0A195F1V2 A0A232FDI7 A0A151XBJ3 E9IF96 A0A0V0G8P1 R4G436 E2B6I9 A0A151JT30 F4WFW0 K7J316 A0A158NL56 E2AI00 A0A195BH09 A0A0L7QTM9 R4WSH1 A0A023FA46 A0A195C023 A0A0P4VIQ4 A0A2P8YKD3 A0A1B6CXA4 T1J971 A0A1W4X7I5 A0A088AB43 A0A1B6KTR5 A0A1W4X7C1 A0A1B6GVV2 A0A1B0CJ92 A0A1Y1MNA9 A0A154P3D7 A0A1L8D928 A0A0M8ZVT1 A0A0K8TM03 D6WT95 U5EYB0 V5HWY2 A0A2P6KEU0 A0A1Z5LEE2 A0A2R5L753 A0A182HA92 A0A023ENZ0 A0A0P4WNI5 A0A182G0Y9 A0A2L2YF39 Q17EV3 A0A1Q3F6L7 A0A210PXI3 B0WYU5 A0A1S3J776 A0A1S3J6X0 A0A1W4X7D6 W5MSE2 A0A182JE75 A0A0K2TXC8 A0A3B5K4H1 D3PIB2 A0A131XCF8 A0A3B4FNW5 I3JZN4 A0A3P8NVW3 A0A3P9CKS1 A0A182JRL3 L7M342 A0A3Q1B8Z3 A0A3P8SEQ6 F1R9P8 Q6DHM8 A0A182PH23 A0A3Q1EKZ0 A0A224Z195 H3CS83 A0A182TP44 C3XZ44 A0A1C7CZT6 A0A2C9GQT7 A0A182ULQ9 Q7PXL1 Q4SMG2 A0A131YPY2 H3BHC8 A0A3N0XVL8 A0A182LTL3 H3BHC9 I3JZN5 S4RFW1 A0A182RF22 S4RFW0
A0A2H1VQM8 A0A212EW10 A0A0A9XNA6 A0A026WZ80 A0A2A3EPZ4 V9IJA0 A0A2R7WCA8 A0A067RTL4 A0A195F1V2 A0A232FDI7 A0A151XBJ3 E9IF96 A0A0V0G8P1 R4G436 E2B6I9 A0A151JT30 F4WFW0 K7J316 A0A158NL56 E2AI00 A0A195BH09 A0A0L7QTM9 R4WSH1 A0A023FA46 A0A195C023 A0A0P4VIQ4 A0A2P8YKD3 A0A1B6CXA4 T1J971 A0A1W4X7I5 A0A088AB43 A0A1B6KTR5 A0A1W4X7C1 A0A1B6GVV2 A0A1B0CJ92 A0A1Y1MNA9 A0A154P3D7 A0A1L8D928 A0A0M8ZVT1 A0A0K8TM03 D6WT95 U5EYB0 V5HWY2 A0A2P6KEU0 A0A1Z5LEE2 A0A2R5L753 A0A182HA92 A0A023ENZ0 A0A0P4WNI5 A0A182G0Y9 A0A2L2YF39 Q17EV3 A0A1Q3F6L7 A0A210PXI3 B0WYU5 A0A1S3J776 A0A1S3J6X0 A0A1W4X7D6 W5MSE2 A0A182JE75 A0A0K2TXC8 A0A3B5K4H1 D3PIB2 A0A131XCF8 A0A3B4FNW5 I3JZN4 A0A3P8NVW3 A0A3P9CKS1 A0A182JRL3 L7M342 A0A3Q1B8Z3 A0A3P8SEQ6 F1R9P8 Q6DHM8 A0A182PH23 A0A3Q1EKZ0 A0A224Z195 H3CS83 A0A182TP44 C3XZ44 A0A1C7CZT6 A0A2C9GQT7 A0A182ULQ9 Q7PXL1 Q4SMG2 A0A131YPY2 H3BHC8 A0A3N0XVL8 A0A182LTL3 H3BHC9 I3JZN5 S4RFW1 A0A182RF22 S4RFW0
Ontologies
PANTHER
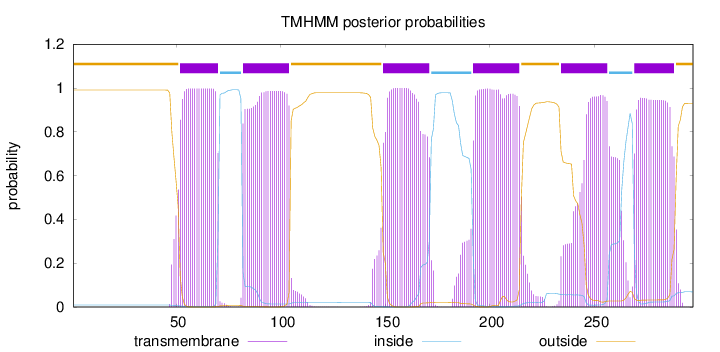
Topology
Length:
297
Number of predicted TMHs:
6
Exp number of AAs in TMHs:
130.72872
Exp number, first 60 AAs:
10.24269
Total prob of N-in:
0.00927
POSSIBLE N-term signal
sequence
outside
1 - 51
TMhelix
52 - 70
inside
71 - 81
TMhelix
82 - 104
outside
105 - 148
TMhelix
149 - 171
inside
172 - 191
TMhelix
192 - 214
outside
215 - 233
TMhelix
234 - 256
inside
257 - 268
TMhelix
269 - 288
outside
289 - 297
Population Genetic Test Statistics
Pi
264.219949
Theta
217.200076
Tajima's D
1.771578
CLR
0.498952
CSRT
0.843857807109645
Interpretation
Uncertain
