Gene
KWMTBOMO15192
Pre Gene Modal
BGIBMGA004945
Annotation
Protein_NDRG3_[Operophtera_brumata]
Location in the cell
Extracellular Reliability : 1.272 PlasmaMembrane Reliability : 1.394
Sequence
CDS
ATGCCGACATCCGTGCACGAAAACGTTGCTCTCCTCAACATGATGTCTTCGGATAATTACGAAGACGATCTCAAAAGTGTTCAACTGCATTTTCCATCCGAAAGACGTTTGCAAAGTGACGGCGCATGCGTGGAGGAGAGGGTGCGGACTTCCCGCGGCGACATCCTCGTTGCCGTACGAGGGGACCGCAGCAAACGAGCTATAATTACTTATCATGACCTTGGACTCAATTATGCCGCCAACTTTCAGGCGTTCTTTAACTTCGTCGATATGAGATCAATTCTGGAACAATTTTGCATCTACCACGTAAACGCTCCTGGCCAAGAAGAGGGATCTCCGACTCTACCCGAGGATTACACCTATCCTAGCATGGACGCTCTGGCGTCTCAGATCGACTTCGTCTTGGGTCATTTTGGTATACGATCTTTCATCGGTTTCGGAGTGGGTGCCGGAGCGAACATATTATCGCGTTATGCCATCATCAACCCCCAAAAAGTAGATGCTTTAGTGCTCATTAACTGCACCTCTACCCAAGCGGGTTGGACCGAGTGGCTGTACCAAAAAATTAATACAAGACAACTGCGCGCTAACGGAATGACTCAAGGCGCGCTGGACTACCTGATGTGGCACCACTTCGGTCGAAGCACGGAGGATCGCAACCATGACCTCGCTCAAGTATACAAAGATTGCTTTTCTCATGTGAACCCTGTCAATTTGTCATTGTTCATCGACAGCTACTTACGGCGCACGGACCTGAGTATAGCGCGCGATTCAAACACACTGAAGGTGCCAGTTCTAAATGTGACCGGCGCTTTGTCGCCTCATGTCGATGATACGGTGACATTTAACGGGCGACTAGATCCCGCTAAGACGTCGTGGTTTAGCATTTCCGATAGTGGAATGGTATTGGAAGAGCAGCCAAGTAAGATAGCTGAGGCGTTCAAGTTGTTTCTCCAGGGCGAAGGGTACTTGAGGTAG
Protein
MPTSVHENVALLNMMSSDNYEDDLKSVQLHFPSERRLQSDGACVEERVRTSRGDILVAVRGDRSKRAIITYHDLGLNYAANFQAFFNFVDMRSILEQFCIYHVNAPGQEEGSPTLPEDYTYPSMDALASQIDFVLGHFGIRSFIGFGVGAGANILSRYAIINPQKVDALVLINCTSTQAGWTEWLYQKINTRQLRANGMTQGALDYLMWHHFGRSTEDRNHDLAQVYKDCFSHVNPVNLSLFIDSYLRRTDLSIARDSNTLKVPVLNVTGALSPHVDDTVTFNGRLDPAKTSWFSISDSGMVLEEQPSKIAEAFKLFLQGEGYLR
Summary
Uniprot
H9J604
A0A2A4K621
A0A2H1V6E7
A0A0L7L547
A0A194RL81
A0A194PGZ1
+ More
A0A212FNA1 A0A151WUB5 A0A195D3F7 A0A151JAK7 A0A195FJN0 D6WIE5 E2CA28 A0A1W4WSX3 A0A3L8E0F2 V9I8H3 V9I7Z6 E9IGE7 A0A1W4WGS2 A0A1W4WRK2 V9I801 A0A195BB09 E2AD05 A0A154NXW5 A0A0C9RD71 A0A0C9QIM9 A0A0C9R5X4 A0A026WXL0 A0A0L7QUG1 A0A310S906 A0A1W4WSW7 V9I6U9 V9I6T0 A0A2J7RJQ1 A0A067RB19 A0A1Y1NF36 V9I8W4 A0A1Y1NDF7 A0A088AS25 A0A2J7RJP6 K7J7N9 U4TSW9 A0A1Y1NCE7 A0A1Y1N9T6 A0A1B6CUF5 A0A1B6DTW4 A0A1B6C6P6 A0A1Q3FV82 A0A1B6HET2 A0A1Q3FV01 A0A1Q3G460 A0A1Q3G405 A0A1B6F6K5 A0A1Q3G421 A0A2C9GQ91 A0A1B6JFE1 A0A1S4H024 A0A1Q3FUT8 A0A1Q3FV09 A0A1B6JTG2 A0A182LK27 A0A1Q3FVA3 A0A1B6J8Q5 A0A1S4GZB7 Q7PHU3 A0A1B6KI75 A0A1B6H2T0 A0A0A9W699 A0A1B0CPV8 A0A084VSY4 A0A212EMP0 A0A182NZ05 A0A1J1II89 A0A182QXK3 A0A1Q3FG31 A0A1Q3FG84 A0A1B6LBZ6 A0A182PD28 A0A0Q9XLJ9 A0A0Q9XHA7 A0A1B6DW05 A0A0A9Y7U1 A0A0Q9XJU2 A0A182Y996 A0A0A9Y2A5 A0A1B6ECJ9 A0A0Q9X8C1 A0A1A9X3V4 A0A2M4AG68 D3TPL7 U5ET09 A0A1B6CF76 A0A1B6DUP9 A0A182K3E6 A0A1D2MFZ8 A0A0Q9W5Y9 A0A1B0BGG3 A0A182M0L8 A0A1B6LPC0 A0A1J1II46
A0A212FNA1 A0A151WUB5 A0A195D3F7 A0A151JAK7 A0A195FJN0 D6WIE5 E2CA28 A0A1W4WSX3 A0A3L8E0F2 V9I8H3 V9I7Z6 E9IGE7 A0A1W4WGS2 A0A1W4WRK2 V9I801 A0A195BB09 E2AD05 A0A154NXW5 A0A0C9RD71 A0A0C9QIM9 A0A0C9R5X4 A0A026WXL0 A0A0L7QUG1 A0A310S906 A0A1W4WSW7 V9I6U9 V9I6T0 A0A2J7RJQ1 A0A067RB19 A0A1Y1NF36 V9I8W4 A0A1Y1NDF7 A0A088AS25 A0A2J7RJP6 K7J7N9 U4TSW9 A0A1Y1NCE7 A0A1Y1N9T6 A0A1B6CUF5 A0A1B6DTW4 A0A1B6C6P6 A0A1Q3FV82 A0A1B6HET2 A0A1Q3FV01 A0A1Q3G460 A0A1Q3G405 A0A1B6F6K5 A0A1Q3G421 A0A2C9GQ91 A0A1B6JFE1 A0A1S4H024 A0A1Q3FUT8 A0A1Q3FV09 A0A1B6JTG2 A0A182LK27 A0A1Q3FVA3 A0A1B6J8Q5 A0A1S4GZB7 Q7PHU3 A0A1B6KI75 A0A1B6H2T0 A0A0A9W699 A0A1B0CPV8 A0A084VSY4 A0A212EMP0 A0A182NZ05 A0A1J1II89 A0A182QXK3 A0A1Q3FG31 A0A1Q3FG84 A0A1B6LBZ6 A0A182PD28 A0A0Q9XLJ9 A0A0Q9XHA7 A0A1B6DW05 A0A0A9Y7U1 A0A0Q9XJU2 A0A182Y996 A0A0A9Y2A5 A0A1B6ECJ9 A0A0Q9X8C1 A0A1A9X3V4 A0A2M4AG68 D3TPL7 U5ET09 A0A1B6CF76 A0A1B6DUP9 A0A182K3E6 A0A1D2MFZ8 A0A0Q9W5Y9 A0A1B0BGG3 A0A182M0L8 A0A1B6LPC0 A0A1J1II46
Pubmed
EMBL
BABH01029906
NWSH01000091
PCG79677.1
ODYU01000905
SOQ36369.1
JTDY01002937
+ More
KOB70456.1 KQ460045 KPJ18179.1 KQ459604 KPI92313.1 AGBW02006247 OWR55224.1 KQ982753 KYQ51225.1 KQ976885 KYN07391.1 KQ979302 KYN21983.1 KQ981523 KYN40209.1 KQ971334 EFA01066.1 GL453920 EFN75183.1 QOIP01000002 RLU25659.1 JR035902 AEY56771.1 JR035898 JR035899 AEY56767.1 AEY56768.1 GL762997 EFZ20356.1 JR035903 AEY56772.1 KQ976530 KYM81718.1 GL438632 EFN68689.1 KQ434772 KZC03934.1 GBYB01004866 GBYB01004876 JAG74633.1 JAG74643.1 GBYB01003364 JAG73131.1 GBYB01003360 JAG73127.1 KK107069 EZA60767.1 KQ414736 KOC62101.1 KQ763659 OAD54893.1 JR035900 JR035901 AEY56769.1 AEY56770.1 JR035904 AEY56773.1 NEVH01002989 PNF41064.1 KK852768 KDR16945.1 GEZM01009844 JAV94267.1 JR035905 AEY56774.1 GEZM01009845 JAV94266.1 PNF41065.1 AAZX01002864 KB631607 ERL84619.1 GEZM01009846 JAV94265.1 GEZM01009843 JAV94268.1 GEDC01020176 JAS17122.1 GEDC01008213 JAS29085.1 GEDC01028224 JAS09074.1 GFDL01003693 JAV31352.1 GECU01034561 JAS73145.1 GFDL01003687 JAV31358.1 GFDL01000458 JAV34587.1 GFDL01000515 JAV34530.1 GECZ01024276 JAS45493.1 GFDL01000498 JAV34547.1 APCN01000634 GECU01009791 GECU01004409 JAS97915.1 JAT03298.1 AAAB01008944 GFDL01003674 JAV31371.1 GFDL01003677 JAV31368.1 GECU01005188 JAT02519.1 GFDL01003673 JAV31372.1 GECU01012142 JAS95564.1 EAA44402.3 GEBQ01028862 JAT11115.1 GECZ01000839 JAS68930.1 GBHO01040673 GBRD01002285 JAG02931.1 JAG63536.1 AJWK01022691 AJWK01022692 AJWK01022693 AJWK01022694 AJWK01022695 ATLV01016150 KE525057 KFB41078.1 AGBW02013811 OWR42739.1 CVRI01000054 CRK99923.1 AXCN02000732 GFDL01008536 JAV26509.1 GFDL01008533 JAV26512.1 GEBQ01018768 JAT21209.1 CH933808 KRG04627.1 KRG04628.1 GEDC01007471 JAS29827.1 GBHO01016441 GDHC01005623 JAG27163.1 JAQ13006.1 KRG04625.1 GBHO01016437 GBRD01015544 JAG27167.1 JAG50282.1 GEDC01001704 JAS35594.1 KRG04623.1 GGFK01006464 MBW39785.1 EZ423369 ADD19645.1 GANO01002990 JAB56881.1 GEDC01025182 GEDC01025069 JAS12116.1 JAS12229.1 GEDC01011597 GEDC01007898 JAS25701.1 JAS29400.1 LJIJ01001385 ODM91893.1 CH940648 KRF80328.1 JXJN01013840 JXJN01013841 AXCM01002314 GEBQ01014520 JAT25457.1 CRK99925.1
KOB70456.1 KQ460045 KPJ18179.1 KQ459604 KPI92313.1 AGBW02006247 OWR55224.1 KQ982753 KYQ51225.1 KQ976885 KYN07391.1 KQ979302 KYN21983.1 KQ981523 KYN40209.1 KQ971334 EFA01066.1 GL453920 EFN75183.1 QOIP01000002 RLU25659.1 JR035902 AEY56771.1 JR035898 JR035899 AEY56767.1 AEY56768.1 GL762997 EFZ20356.1 JR035903 AEY56772.1 KQ976530 KYM81718.1 GL438632 EFN68689.1 KQ434772 KZC03934.1 GBYB01004866 GBYB01004876 JAG74633.1 JAG74643.1 GBYB01003364 JAG73131.1 GBYB01003360 JAG73127.1 KK107069 EZA60767.1 KQ414736 KOC62101.1 KQ763659 OAD54893.1 JR035900 JR035901 AEY56769.1 AEY56770.1 JR035904 AEY56773.1 NEVH01002989 PNF41064.1 KK852768 KDR16945.1 GEZM01009844 JAV94267.1 JR035905 AEY56774.1 GEZM01009845 JAV94266.1 PNF41065.1 AAZX01002864 KB631607 ERL84619.1 GEZM01009846 JAV94265.1 GEZM01009843 JAV94268.1 GEDC01020176 JAS17122.1 GEDC01008213 JAS29085.1 GEDC01028224 JAS09074.1 GFDL01003693 JAV31352.1 GECU01034561 JAS73145.1 GFDL01003687 JAV31358.1 GFDL01000458 JAV34587.1 GFDL01000515 JAV34530.1 GECZ01024276 JAS45493.1 GFDL01000498 JAV34547.1 APCN01000634 GECU01009791 GECU01004409 JAS97915.1 JAT03298.1 AAAB01008944 GFDL01003674 JAV31371.1 GFDL01003677 JAV31368.1 GECU01005188 JAT02519.1 GFDL01003673 JAV31372.1 GECU01012142 JAS95564.1 EAA44402.3 GEBQ01028862 JAT11115.1 GECZ01000839 JAS68930.1 GBHO01040673 GBRD01002285 JAG02931.1 JAG63536.1 AJWK01022691 AJWK01022692 AJWK01022693 AJWK01022694 AJWK01022695 ATLV01016150 KE525057 KFB41078.1 AGBW02013811 OWR42739.1 CVRI01000054 CRK99923.1 AXCN02000732 GFDL01008536 JAV26509.1 GFDL01008533 JAV26512.1 GEBQ01018768 JAT21209.1 CH933808 KRG04627.1 KRG04628.1 GEDC01007471 JAS29827.1 GBHO01016441 GDHC01005623 JAG27163.1 JAQ13006.1 KRG04625.1 GBHO01016437 GBRD01015544 JAG27167.1 JAG50282.1 GEDC01001704 JAS35594.1 KRG04623.1 GGFK01006464 MBW39785.1 EZ423369 ADD19645.1 GANO01002990 JAB56881.1 GEDC01025182 GEDC01025069 JAS12116.1 JAS12229.1 GEDC01011597 GEDC01007898 JAS25701.1 JAS29400.1 LJIJ01001385 ODM91893.1 CH940648 KRF80328.1 JXJN01013840 JXJN01013841 AXCM01002314 GEBQ01014520 JAT25457.1 CRK99925.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000053240
UP000053268
UP000007151
+ More
UP000075809 UP000078542 UP000078492 UP000078541 UP000007266 UP000008237 UP000192223 UP000279307 UP000078540 UP000000311 UP000076502 UP000053097 UP000053825 UP000235965 UP000027135 UP000005203 UP000002358 UP000030742 UP000075840 UP000075882 UP000007062 UP000092461 UP000030765 UP000075884 UP000183832 UP000075886 UP000075885 UP000009192 UP000076408 UP000091820 UP000075881 UP000094527 UP000008792 UP000092460 UP000075883
UP000075809 UP000078542 UP000078492 UP000078541 UP000007266 UP000008237 UP000192223 UP000279307 UP000078540 UP000000311 UP000076502 UP000053097 UP000053825 UP000235965 UP000027135 UP000005203 UP000002358 UP000030742 UP000075840 UP000075882 UP000007062 UP000092461 UP000030765 UP000075884 UP000183832 UP000075886 UP000075885 UP000009192 UP000076408 UP000091820 UP000075881 UP000094527 UP000008792 UP000092460 UP000075883
PRIDE
Pfam
PF03096 Ndr
SUPFAM
SSF53474
SSF53474
Gene 3D
ProteinModelPortal
H9J604
A0A2A4K621
A0A2H1V6E7
A0A0L7L547
A0A194RL81
A0A194PGZ1
+ More
A0A212FNA1 A0A151WUB5 A0A195D3F7 A0A151JAK7 A0A195FJN0 D6WIE5 E2CA28 A0A1W4WSX3 A0A3L8E0F2 V9I8H3 V9I7Z6 E9IGE7 A0A1W4WGS2 A0A1W4WRK2 V9I801 A0A195BB09 E2AD05 A0A154NXW5 A0A0C9RD71 A0A0C9QIM9 A0A0C9R5X4 A0A026WXL0 A0A0L7QUG1 A0A310S906 A0A1W4WSW7 V9I6U9 V9I6T0 A0A2J7RJQ1 A0A067RB19 A0A1Y1NF36 V9I8W4 A0A1Y1NDF7 A0A088AS25 A0A2J7RJP6 K7J7N9 U4TSW9 A0A1Y1NCE7 A0A1Y1N9T6 A0A1B6CUF5 A0A1B6DTW4 A0A1B6C6P6 A0A1Q3FV82 A0A1B6HET2 A0A1Q3FV01 A0A1Q3G460 A0A1Q3G405 A0A1B6F6K5 A0A1Q3G421 A0A2C9GQ91 A0A1B6JFE1 A0A1S4H024 A0A1Q3FUT8 A0A1Q3FV09 A0A1B6JTG2 A0A182LK27 A0A1Q3FVA3 A0A1B6J8Q5 A0A1S4GZB7 Q7PHU3 A0A1B6KI75 A0A1B6H2T0 A0A0A9W699 A0A1B0CPV8 A0A084VSY4 A0A212EMP0 A0A182NZ05 A0A1J1II89 A0A182QXK3 A0A1Q3FG31 A0A1Q3FG84 A0A1B6LBZ6 A0A182PD28 A0A0Q9XLJ9 A0A0Q9XHA7 A0A1B6DW05 A0A0A9Y7U1 A0A0Q9XJU2 A0A182Y996 A0A0A9Y2A5 A0A1B6ECJ9 A0A0Q9X8C1 A0A1A9X3V4 A0A2M4AG68 D3TPL7 U5ET09 A0A1B6CF76 A0A1B6DUP9 A0A182K3E6 A0A1D2MFZ8 A0A0Q9W5Y9 A0A1B0BGG3 A0A182M0L8 A0A1B6LPC0 A0A1J1II46
A0A212FNA1 A0A151WUB5 A0A195D3F7 A0A151JAK7 A0A195FJN0 D6WIE5 E2CA28 A0A1W4WSX3 A0A3L8E0F2 V9I8H3 V9I7Z6 E9IGE7 A0A1W4WGS2 A0A1W4WRK2 V9I801 A0A195BB09 E2AD05 A0A154NXW5 A0A0C9RD71 A0A0C9QIM9 A0A0C9R5X4 A0A026WXL0 A0A0L7QUG1 A0A310S906 A0A1W4WSW7 V9I6U9 V9I6T0 A0A2J7RJQ1 A0A067RB19 A0A1Y1NF36 V9I8W4 A0A1Y1NDF7 A0A088AS25 A0A2J7RJP6 K7J7N9 U4TSW9 A0A1Y1NCE7 A0A1Y1N9T6 A0A1B6CUF5 A0A1B6DTW4 A0A1B6C6P6 A0A1Q3FV82 A0A1B6HET2 A0A1Q3FV01 A0A1Q3G460 A0A1Q3G405 A0A1B6F6K5 A0A1Q3G421 A0A2C9GQ91 A0A1B6JFE1 A0A1S4H024 A0A1Q3FUT8 A0A1Q3FV09 A0A1B6JTG2 A0A182LK27 A0A1Q3FVA3 A0A1B6J8Q5 A0A1S4GZB7 Q7PHU3 A0A1B6KI75 A0A1B6H2T0 A0A0A9W699 A0A1B0CPV8 A0A084VSY4 A0A212EMP0 A0A182NZ05 A0A1J1II89 A0A182QXK3 A0A1Q3FG31 A0A1Q3FG84 A0A1B6LBZ6 A0A182PD28 A0A0Q9XLJ9 A0A0Q9XHA7 A0A1B6DW05 A0A0A9Y7U1 A0A0Q9XJU2 A0A182Y996 A0A0A9Y2A5 A0A1B6ECJ9 A0A0Q9X8C1 A0A1A9X3V4 A0A2M4AG68 D3TPL7 U5ET09 A0A1B6CF76 A0A1B6DUP9 A0A182K3E6 A0A1D2MFZ8 A0A0Q9W5Y9 A0A1B0BGG3 A0A182M0L8 A0A1B6LPC0 A0A1J1II46
PDB
2XMR
E-value=4.43395e-50,
Score=499
Ontologies
PANTHER
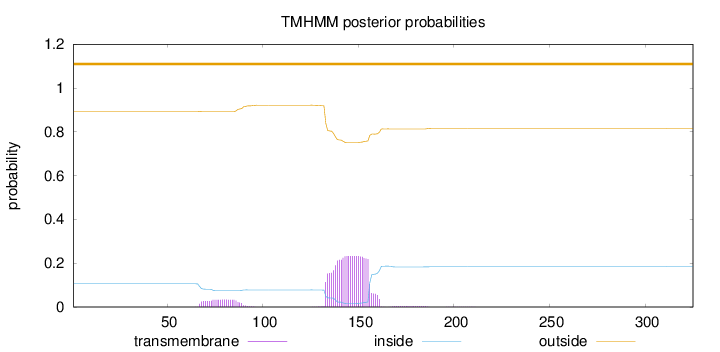
Topology
Length:
325
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
5.95013
Exp number, first 60 AAs:
0
Total prob of N-in:
0.10583
outside
1 - 325
Population Genetic Test Statistics
Pi
308.307019
Theta
218.276002
Tajima's D
1.25352
CLR
0.000225
CSRT
0.724963751812409
Interpretation
Uncertain
