Gene
KWMTBOMO15184
Pre Gene Modal
BGIBMGA004971
Annotation
PREDICTED:_exocyst_complex_component_5_[Plutella_xylostella]
Full name
Exocyst complex component 5
Alternative Name
Exocyst complex component Sec10
Location in the cell
Cytoplasmic Reliability : 1.359 Golgi Reliability : 1.008 Nuclear Reliability : 1.097
Sequence
CDS
ATGCTCAACGATGCTAAAACGTCTTTCCTTAGATGTAAAACGCTGTGTACGCCCAATGAATTGCCGCACACAGCCATGACTCTACTGGAGGTGGTCACGCAACATTTACTGATAGAACACGTTGATTATGCATTGGATTTGGGTCTGCAGTCTATACCTATCGCGGAAGGCAAGTCGCCACCTCAGATATATTTCTTTGATACTGTAAAACAAGCTAATGTAATTGTGAGAATGTTCGAGGAACATTTCCAGGAGTCGATTTTACCTTGTATAAGCTCATCGTCGAAACAAACAGAATGTTTACACAAGAAGACCGCTATAATGGACCAACTCGAGAGCAAACTTGACGCTGGCTTGGAACGTGCTATATCGGCTATAGTGAGCTGGGTGAAGCTGCATCTGCAGACCGAACAAAAGAAGACTGACTTCAAACCGGAAGGGGACATCGATACTATAGCGTCACCAGCTTGCCTAGCAGTGGTGTCCTATTTGAATTCGGTAATGGATAAAATACACGATGGTTTGGAAGACGACAATAGAAATTCCATCTCGTTGGAGCTGGCCGTGAGACTGCACAGGGTTATATACGAACACCTGCATAACTTCCAGTTTAATTCGGCAGGTGCTATGATAGCGATATGCGACGTGAAGGAGTACAGGTCGGCCGTGTGCGGGCGCGGCTCGAGGGCGGCTCCGGCTCCGGCCCACGGGCTGTTCGACGCTCTGCACGCGCTCTGCAATCTGCTGCTCGTCAAACCGGAGAACCTGCATCAGGTTTGCGAGGGAGAGACCCTGGCGGAGTTTGATCATTCGATTCTGTTGAGCTTCATACAGCTCCGAGCGGATTACAAGAGTCACAAGCTATCCACCTTCCTCAAGGGACTGTCTTAA
Protein
MLNDAKTSFLRCKTLCTPNELPHTAMTLLEVVTQHLLIEHVDYALDLGLQSIPIAEGKSPPQIYFFDTVKQANVIVRMFEEHFQESILPCISSSSKQTECLHKKTAIMDQLESKLDAGLERAISAIVSWVKLHLQTEQKKTDFKPEGDIDTIASPACLAVVSYLNSVMDKIHDGLEDDNRNSISLELAVRLHRVIYEHLHNFQFNSAGAMIAICDVKEYRSAVCGRGSRAAPAPAHGLFDALHALCNLLLVKPENLHQVCEGETLAEFDHSILLSFIQLRADYKSHKLSTFLKGLS
Summary
Subunit
The exocyst complex is composed of Sec3/Exoc1, Sec5/Exoc2, Sec6/Exoc3, Sec8/Exoc4, Sec10/Exoc5, Sec15/Exoc6, Exo70/Exoc7 and Exo84/Exoc8.
Similarity
Belongs to the SEC10 family.
Keywords
Coiled coil
Complete proteome
Exocytosis
Protein transport
Reference proteome
Transport
Feature
chain Exocyst complex component 5
Uniprot
A0A3S2N656
A0A2H1WJR2
A0A194PGY6
A0A212ENJ3
A0A2A4K627
A0A194RK24
+ More
H9J630 A0A1E1WJN8 A0A1E1WMK5 S4PXN6 A0A2J7QLM0 A0A067QT50 A0A154PHG7 A0A0L7QMF3 A0A087ZT75 E2A2D2 A0A0M9A6T0 E2BZ07 E0VIG2 A0A0J7L003 A0A1B0CC55 A0A195EKP9 E9J266 A0A1B0DG62 K7JB79 A0A195FSM2 A0A232F0A7 A0A195CUS6 A0A151WWK7 A0A026WJS9 A0A1B6HK07 A0A158NA17 A0A2A3EBN6 A0A195BKA7 F4WC45 A0A1B6CAB8 A0A1B6FMR3 A0A1B6KSQ6 A0A1B6D928 A0A0V0G5G3 A0A0M4EN18 A0A069DWN9 A0A182S7M2 B4JIG3 A0A1J1HHS2 A0A0K8V0G5 B4G417 A0A182G579 B4K7E1 B5DVK9 B4MBY6 A0A3B0KCH6 S5MHR4 Q9XTM1 A0A1W4V6K5 B4PL16 B4HGI3 A0A1S4F9F4 B0WQE8 Q17B75 A0A182JPR0 B3M0L9 A0A084WTM7 B3P791 A0A1J1HPY2 A0A182ILN1 B4NBM9 A0A182XJ12 A0A182UPM1 A0A182RYL4 A0A0P4VYV2 A0A182W1J9 A0A0A1X774 A0A182L7E8 Q7PQN8 A0A182I7U3 W8BCQ8 A0A182MQ72 A0A1Q3FCE7 A0A182YGL5 A0A182TYR9 A0A182PPY7 W5J463 A0A182QVJ4 K1Q2T9 D6WU95 A0A034W4P0 A0A182FPJ2 A0A0K8SSN0 R7V2S8 A0A182N5R4 A0A2M4AK51 E9HYN5 A0A1Y1MPN6 A0A3Q0ITS5 A0A0A9W0K1 A0A1A9X184 A0A0A9XDW3 A0A0N8BDI3 A0A210QYB7
H9J630 A0A1E1WJN8 A0A1E1WMK5 S4PXN6 A0A2J7QLM0 A0A067QT50 A0A154PHG7 A0A0L7QMF3 A0A087ZT75 E2A2D2 A0A0M9A6T0 E2BZ07 E0VIG2 A0A0J7L003 A0A1B0CC55 A0A195EKP9 E9J266 A0A1B0DG62 K7JB79 A0A195FSM2 A0A232F0A7 A0A195CUS6 A0A151WWK7 A0A026WJS9 A0A1B6HK07 A0A158NA17 A0A2A3EBN6 A0A195BKA7 F4WC45 A0A1B6CAB8 A0A1B6FMR3 A0A1B6KSQ6 A0A1B6D928 A0A0V0G5G3 A0A0M4EN18 A0A069DWN9 A0A182S7M2 B4JIG3 A0A1J1HHS2 A0A0K8V0G5 B4G417 A0A182G579 B4K7E1 B5DVK9 B4MBY6 A0A3B0KCH6 S5MHR4 Q9XTM1 A0A1W4V6K5 B4PL16 B4HGI3 A0A1S4F9F4 B0WQE8 Q17B75 A0A182JPR0 B3M0L9 A0A084WTM7 B3P791 A0A1J1HPY2 A0A182ILN1 B4NBM9 A0A182XJ12 A0A182UPM1 A0A182RYL4 A0A0P4VYV2 A0A182W1J9 A0A0A1X774 A0A182L7E8 Q7PQN8 A0A182I7U3 W8BCQ8 A0A182MQ72 A0A1Q3FCE7 A0A182YGL5 A0A182TYR9 A0A182PPY7 W5J463 A0A182QVJ4 K1Q2T9 D6WU95 A0A034W4P0 A0A182FPJ2 A0A0K8SSN0 R7V2S8 A0A182N5R4 A0A2M4AK51 E9HYN5 A0A1Y1MPN6 A0A3Q0ITS5 A0A0A9W0K1 A0A1A9X184 A0A0A9XDW3 A0A0N8BDI3 A0A210QYB7
Pubmed
26354079
22118469
19121390
23622113
24845553
20798317
+ More
20566863 21282665 20075255 28648823 24508170 30249741 21347285 21719571 26334808 17994087 26483478 15632085 10731132 12537572 12537569 17550304 17510324 24438588 27129103 25830018 20966253 12364791 14747013 17210077 24495485 25244985 20920257 23761445 22992520 18362917 19820115 25348373 23254933 21292972 28004739 25401762 28812685
20566863 21282665 20075255 28648823 24508170 30249741 21347285 21719571 26334808 17994087 26483478 15632085 10731132 12537572 12537569 17550304 17510324 24438588 27129103 25830018 20966253 12364791 14747013 17210077 24495485 25244985 20920257 23761445 22992520 18362917 19820115 25348373 23254933 21292972 28004739 25401762 28812685
EMBL
RSAL01000329
RVE42509.1
ODYU01009039
SOQ53152.1
KQ459604
KPI92308.1
+ More
AGBW02013634 OWR43062.1 NWSH01000091 PCG79687.1 KQ460045 KPJ18183.1 BABH01029921 BABH01029922 GDQN01003831 JAT87223.1 GDQN01002801 JAT88253.1 GAIX01004723 JAA87837.1 NEVH01013243 PNF29480.1 KK853567 KDR06665.1 KQ434905 KZC11283.1 KQ414894 KOC59711.1 GL436037 EFN72395.1 KQ435736 KOX77029.1 GL451538 EFN79079.1 DS235199 EEB13168.1 LBMM01001666 KMQ95923.1 AJWK01006156 KQ978739 KYN28716.1 GL767819 EFZ13092.1 AJVK01059305 KQ981305 KYN42899.1 NNAY01001403 OXU24091.1 KQ977276 KYN04436.1 KQ982686 KYQ52294.1 KK107182 QOIP01000011 EZA55921.1 RLU16558.1 GECU01032714 JAS74992.1 ADTU01009939 KZ288292 PBC29100.1 KQ976455 KYM85083.1 GL888070 EGI68025.1 GEDC01026866 JAS10432.1 GECZ01018271 JAS51498.1 GEBQ01026844 GEBQ01025522 GEBQ01022292 JAT13133.1 JAT14455.1 JAT17685.1 GEDC01015115 JAS22183.1 GECL01002833 JAP03291.1 CP012526 ALC45976.1 GBGD01000638 JAC88251.1 CH916369 EDV93044.1 CVRI01000002 CRK86964.1 GDHF01020041 GDHF01011623 JAI32273.1 JAI40691.1 CH479179 EDW24428.1 JXUM01042954 JXUM01044211 KQ561391 KQ561344 KXJ78726.1 KXJ78892.1 CH933806 EDW14265.1 CM000070 EDY68045.1 CH940656 EDW58607.1 OUUW01000007 SPP82711.1 BT150181 AGR50375.1 AJ242993 AJ242992 AE014297 AY058414 CM000160 EDW99001.1 CH480815 EDW43436.1 DS232039 EDS32819.1 CH477325 EAT43492.1 CH902617 EDV44266.1 ATLV01026902 KE525420 KFB53571.1 CH954182 EDV53911.1 CVRI01000015 CRK90027.1 CH964232 EDW81193.1 GDKW01000800 JAI55795.1 GBXI01007566 GBXI01003177 JAD06726.1 JAD11115.1 AAAB01008880 EAA08562.4 APCN01002556 GAMC01007505 JAB99050.1 AXCM01000330 GFDL01009872 JAV25173.1 ADMH02002100 ETN59197.1 AXCN02001158 JH816014 EKC23175.1 KQ971352 EFA06250.1 GAKP01010253 GAKP01010251 GAKP01010249 GAKP01010247 JAC48703.1 GBRD01009645 JAG56179.1 AMQN01005993 KB297336 ELU10626.1 GGFK01007844 MBW41165.1 GL733196 EFX63140.1 GEZM01029807 GEZM01029806 JAV85916.1 GBHO01042240 JAG01364.1 GBHO01025450 JAG18154.1 GDIQ01188395 JAK63330.1 NEDP02001218 OWF53753.1
AGBW02013634 OWR43062.1 NWSH01000091 PCG79687.1 KQ460045 KPJ18183.1 BABH01029921 BABH01029922 GDQN01003831 JAT87223.1 GDQN01002801 JAT88253.1 GAIX01004723 JAA87837.1 NEVH01013243 PNF29480.1 KK853567 KDR06665.1 KQ434905 KZC11283.1 KQ414894 KOC59711.1 GL436037 EFN72395.1 KQ435736 KOX77029.1 GL451538 EFN79079.1 DS235199 EEB13168.1 LBMM01001666 KMQ95923.1 AJWK01006156 KQ978739 KYN28716.1 GL767819 EFZ13092.1 AJVK01059305 KQ981305 KYN42899.1 NNAY01001403 OXU24091.1 KQ977276 KYN04436.1 KQ982686 KYQ52294.1 KK107182 QOIP01000011 EZA55921.1 RLU16558.1 GECU01032714 JAS74992.1 ADTU01009939 KZ288292 PBC29100.1 KQ976455 KYM85083.1 GL888070 EGI68025.1 GEDC01026866 JAS10432.1 GECZ01018271 JAS51498.1 GEBQ01026844 GEBQ01025522 GEBQ01022292 JAT13133.1 JAT14455.1 JAT17685.1 GEDC01015115 JAS22183.1 GECL01002833 JAP03291.1 CP012526 ALC45976.1 GBGD01000638 JAC88251.1 CH916369 EDV93044.1 CVRI01000002 CRK86964.1 GDHF01020041 GDHF01011623 JAI32273.1 JAI40691.1 CH479179 EDW24428.1 JXUM01042954 JXUM01044211 KQ561391 KQ561344 KXJ78726.1 KXJ78892.1 CH933806 EDW14265.1 CM000070 EDY68045.1 CH940656 EDW58607.1 OUUW01000007 SPP82711.1 BT150181 AGR50375.1 AJ242993 AJ242992 AE014297 AY058414 CM000160 EDW99001.1 CH480815 EDW43436.1 DS232039 EDS32819.1 CH477325 EAT43492.1 CH902617 EDV44266.1 ATLV01026902 KE525420 KFB53571.1 CH954182 EDV53911.1 CVRI01000015 CRK90027.1 CH964232 EDW81193.1 GDKW01000800 JAI55795.1 GBXI01007566 GBXI01003177 JAD06726.1 JAD11115.1 AAAB01008880 EAA08562.4 APCN01002556 GAMC01007505 JAB99050.1 AXCM01000330 GFDL01009872 JAV25173.1 ADMH02002100 ETN59197.1 AXCN02001158 JH816014 EKC23175.1 KQ971352 EFA06250.1 GAKP01010253 GAKP01010251 GAKP01010249 GAKP01010247 JAC48703.1 GBRD01009645 JAG56179.1 AMQN01005993 KB297336 ELU10626.1 GGFK01007844 MBW41165.1 GL733196 EFX63140.1 GEZM01029807 GEZM01029806 JAV85916.1 GBHO01042240 JAG01364.1 GBHO01025450 JAG18154.1 GDIQ01188395 JAK63330.1 NEDP02001218 OWF53753.1
Proteomes
UP000283053
UP000053268
UP000007151
UP000218220
UP000053240
UP000005204
+ More
UP000235965 UP000027135 UP000076502 UP000053825 UP000005203 UP000000311 UP000053105 UP000008237 UP000009046 UP000036403 UP000092461 UP000078492 UP000092462 UP000002358 UP000078541 UP000215335 UP000078542 UP000075809 UP000053097 UP000279307 UP000005205 UP000242457 UP000078540 UP000007755 UP000092553 UP000075901 UP000001070 UP000183832 UP000008744 UP000069940 UP000249989 UP000009192 UP000001819 UP000008792 UP000268350 UP000000803 UP000192221 UP000002282 UP000001292 UP000002320 UP000008820 UP000075881 UP000007801 UP000030765 UP000008711 UP000075880 UP000007798 UP000076407 UP000075903 UP000075900 UP000075920 UP000075882 UP000007062 UP000075840 UP000075883 UP000076408 UP000075902 UP000075885 UP000000673 UP000075886 UP000005408 UP000007266 UP000069272 UP000014760 UP000075884 UP000000305 UP000079169 UP000091820 UP000242188
UP000235965 UP000027135 UP000076502 UP000053825 UP000005203 UP000000311 UP000053105 UP000008237 UP000009046 UP000036403 UP000092461 UP000078492 UP000092462 UP000002358 UP000078541 UP000215335 UP000078542 UP000075809 UP000053097 UP000279307 UP000005205 UP000242457 UP000078540 UP000007755 UP000092553 UP000075901 UP000001070 UP000183832 UP000008744 UP000069940 UP000249989 UP000009192 UP000001819 UP000008792 UP000268350 UP000000803 UP000192221 UP000002282 UP000001292 UP000002320 UP000008820 UP000075881 UP000007801 UP000030765 UP000008711 UP000075880 UP000007798 UP000076407 UP000075903 UP000075900 UP000075920 UP000075882 UP000007062 UP000075840 UP000075883 UP000076408 UP000075902 UP000075885 UP000000673 UP000075886 UP000005408 UP000007266 UP000069272 UP000014760 UP000075884 UP000000305 UP000079169 UP000091820 UP000242188
Interpro
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
A0A3S2N656
A0A2H1WJR2
A0A194PGY6
A0A212ENJ3
A0A2A4K627
A0A194RK24
+ More
H9J630 A0A1E1WJN8 A0A1E1WMK5 S4PXN6 A0A2J7QLM0 A0A067QT50 A0A154PHG7 A0A0L7QMF3 A0A087ZT75 E2A2D2 A0A0M9A6T0 E2BZ07 E0VIG2 A0A0J7L003 A0A1B0CC55 A0A195EKP9 E9J266 A0A1B0DG62 K7JB79 A0A195FSM2 A0A232F0A7 A0A195CUS6 A0A151WWK7 A0A026WJS9 A0A1B6HK07 A0A158NA17 A0A2A3EBN6 A0A195BKA7 F4WC45 A0A1B6CAB8 A0A1B6FMR3 A0A1B6KSQ6 A0A1B6D928 A0A0V0G5G3 A0A0M4EN18 A0A069DWN9 A0A182S7M2 B4JIG3 A0A1J1HHS2 A0A0K8V0G5 B4G417 A0A182G579 B4K7E1 B5DVK9 B4MBY6 A0A3B0KCH6 S5MHR4 Q9XTM1 A0A1W4V6K5 B4PL16 B4HGI3 A0A1S4F9F4 B0WQE8 Q17B75 A0A182JPR0 B3M0L9 A0A084WTM7 B3P791 A0A1J1HPY2 A0A182ILN1 B4NBM9 A0A182XJ12 A0A182UPM1 A0A182RYL4 A0A0P4VYV2 A0A182W1J9 A0A0A1X774 A0A182L7E8 Q7PQN8 A0A182I7U3 W8BCQ8 A0A182MQ72 A0A1Q3FCE7 A0A182YGL5 A0A182TYR9 A0A182PPY7 W5J463 A0A182QVJ4 K1Q2T9 D6WU95 A0A034W4P0 A0A182FPJ2 A0A0K8SSN0 R7V2S8 A0A182N5R4 A0A2M4AK51 E9HYN5 A0A1Y1MPN6 A0A3Q0ITS5 A0A0A9W0K1 A0A1A9X184 A0A0A9XDW3 A0A0N8BDI3 A0A210QYB7
H9J630 A0A1E1WJN8 A0A1E1WMK5 S4PXN6 A0A2J7QLM0 A0A067QT50 A0A154PHG7 A0A0L7QMF3 A0A087ZT75 E2A2D2 A0A0M9A6T0 E2BZ07 E0VIG2 A0A0J7L003 A0A1B0CC55 A0A195EKP9 E9J266 A0A1B0DG62 K7JB79 A0A195FSM2 A0A232F0A7 A0A195CUS6 A0A151WWK7 A0A026WJS9 A0A1B6HK07 A0A158NA17 A0A2A3EBN6 A0A195BKA7 F4WC45 A0A1B6CAB8 A0A1B6FMR3 A0A1B6KSQ6 A0A1B6D928 A0A0V0G5G3 A0A0M4EN18 A0A069DWN9 A0A182S7M2 B4JIG3 A0A1J1HHS2 A0A0K8V0G5 B4G417 A0A182G579 B4K7E1 B5DVK9 B4MBY6 A0A3B0KCH6 S5MHR4 Q9XTM1 A0A1W4V6K5 B4PL16 B4HGI3 A0A1S4F9F4 B0WQE8 Q17B75 A0A182JPR0 B3M0L9 A0A084WTM7 B3P791 A0A1J1HPY2 A0A182ILN1 B4NBM9 A0A182XJ12 A0A182UPM1 A0A182RYL4 A0A0P4VYV2 A0A182W1J9 A0A0A1X774 A0A182L7E8 Q7PQN8 A0A182I7U3 W8BCQ8 A0A182MQ72 A0A1Q3FCE7 A0A182YGL5 A0A182TYR9 A0A182PPY7 W5J463 A0A182QVJ4 K1Q2T9 D6WU95 A0A034W4P0 A0A182FPJ2 A0A0K8SSN0 R7V2S8 A0A182N5R4 A0A2M4AK51 E9HYN5 A0A1Y1MPN6 A0A3Q0ITS5 A0A0A9W0K1 A0A1A9X184 A0A0A9XDW3 A0A0N8BDI3 A0A210QYB7
PDB
5H11
E-value=9.80773e-69,
Score=659
Ontologies
GO
PANTHER
Topology
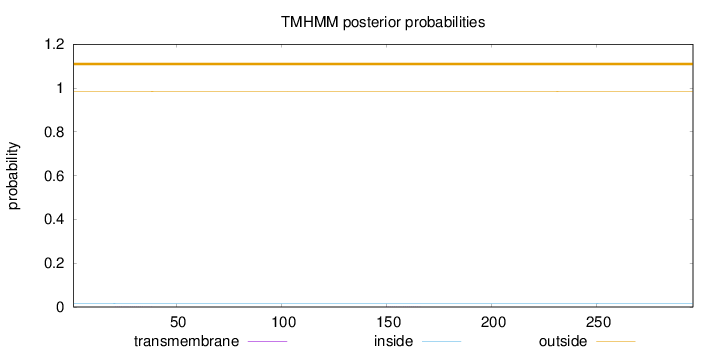
Length:
296
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01057
Exp number, first 60 AAs:
0.0047
Total prob of N-in:
0.01639
outside
1 - 296
Population Genetic Test Statistics
Pi
246.245313
Theta
146.364783
Tajima's D
2.328162
CLR
0.267053
CSRT
0.924403779811009
Interpretation
Uncertain
