Gene
KWMTBOMO15176 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA004939
Annotation
PREDICTED:_pre-mRNA-processing_factor_39_isoform_X1_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 3.429
Sequence
CDS
ATGGAAGATGAAACAAGCTCCGATAATCAGAAAGGATCTGAAAATCCCATGGATACAGACGAAGGTCCTTTGATAAATGAGAACTCCAACGGACTTTCTCATTTGTTAACAGAGGGCGAATTTAATAGTGTCGATGCTGCTGTTGATGAGTCGTCGACATCTAATCAATGTTTCTTGAACGCCGTGGAATTATCAGCTGGAAGCGCTGGTGACTACCTCGAAAACAACGGCCCTCGCTTCAGTACAGATTCTTTTGACATTGGTGATAATGCTGTAACTTTTCCCAATGATTCTCTAAATAGCGCATTGACTGAAGATAATTCTATTCAAAGTGACTCTGCATCCCACAAAGCAGTTTTAGACACAGACAGTAGGCAGTCTGAAAGTCAACATGCAACAGATTCCAATGTAGGAGAGATAAATGAAAACTCTGGCAATGACTCTAACTTCGCCACTGAGCATGTAAATTTAAATGTGGATAAATCAGAGCAATTGGAAATTGTTGAGCCATGTAGCAGTCCAGTAAAAGATCCAGTGATTAAGCAGGATGAAGGCAAAGAGTTTAAAAACCAAAGAGAAGTAGATGATATCAAAGAACAATCACAAGACTCAGCAAGTAAGATTGGTGGTGCTGAAGAAACAGAAGTAGTATCAGAGGATGAACTTCCTGAAGTGCAAAAGCCGAGCATCAAAGATGCTGAGAATGTATCTGATGATGAGTTGCCGGGGCCGAAACCTGCAGAACTGCCTGCTGATACAGAGGTTGTATCAGAAGATGAGCTACCTGCATCCAAGAAAGAGTCTAGAAAACGAAAACCAGATGAAGAGAGAAGTAATTATGATCCAGGATCACCAACATCAGAAGGTGAATCTTCCACAAAAAAACAAGCTATTACCAAGAACGGCGAAAGCAAGCCCATACCTGCCGAAAAGAGACCTTCAGTTGATGAAAAACCTAAAAAGAAAGTTTTACCGGAATTGGAAAAGTACTGGAAAGTTGTGAATGATGATCCGACTGACTTCACTGGGTGGACCTACCTATTACAATATGTCGATCAAGAGAGCGATGTTGAAGCTGCACGTGAAGCCTATGACGCCTTTTTATCCCACTATCCTTATTGCTATGGATATTGGAGAAAGTTTGCAGATTATGAAAAACGTAAAGGAAGTAAAAAGAAGTGCTTAGAGGTTTTGGAAAGGGGACTTAAAGCTATACCTTTATCCGTAGACCTCTGGATACATTACCTCAATCATATAAAAGCGACTAAAACCGAAGACCATGCGTACATCAGATCTCAGTATGAACGAGCCATAGATGCATGCGGACTAGAATTTCGCTCGGATCGTCTTTGGGAGTCTTACATCAAATGGGAAGCGGAAAATGGTTCCGCTCTCAATGTCACGAATATCTACGATCGTCTTTTGGCAACACCCACCTTAGGGTACACGTCACACTTCGACAATTTCCAGGAGCACGTGATGTCGGAGCCGGTGACCGGCGTGGTGAGCCACGCGGAGCTGGTGCGGCTGCGGGGCGAGGTGCGGGACGCGGCCGGCGCGCAGCCCAGCCTCGACCTGCCGCCCGGGGAGGACGAGCCCGTCGACCATGTCGCATCCGATGAAGAGGCGCAAGCCATCAAGGAACGGATTGTTGCAGCTCGAAGAAAAATTCATAAAACGACCGGCGAAGAAGTAACTGCGAGGTGGACTTTCGAAGAAGGAATCAAGCGGCCTTATTTCCACGTGAAGCCCCTGGAACGTTGCCAGCTAAAGAACTGGAAGGCGTACCTCGAATGGGAGAAGCACCACGGTTCGTTCAAGCGAGCCCTCGTGCTCCACGAGAGATGCCTCATTGCATGTGCCTTATATGAAGAATTCTGGATGAAGCTTATAAAGTTTCTGGAAGAACGTGCTGAACAAGATCCTGAAGTGATAGCAATAGAACGGGAAGTACTAGAGCGAGCCTGCACAGTGCATCACCTCGACAAGCCAGAGCTCCACCTACACTGGGCACACTTCGAAGAGGTGCAGGGCAATACACTGAAAGCTGCCGAGATACTTGATCGTATAGAGAAAACCTGTCCCAATCTCGTACAAATACAGTACAGGCGAGTGAACCTCGAGCGACGACGCGGCGACTACGAGAAGTGCTCTCAGCTATACGAAACGTACATCAGCACTTCGAAGAACAAGTCGCTGGCGTCTGCGCTCGCCATCAAGTACGCGCGGTTCCTGCAGCACGTGCGCCGCCAGCCCGCCGCCGCCCGCGCCGCGCTCGACGACGCCGTCGCCAAGGACCCGCTCAACCCGCGCCTGCACCTGCAGAGACTTGACCTCGCCATCAACACTCCCGGCGTCACCTACGAAGAGTTGGATGAGCTGGCACAGTCGTACTCGAGACAGGAGGGCGCGGACACGGAGGTGAGCGCGCACATGGCGTGGCGCCGCCGCGAGCTCGCGGAGGAGCTGGGCAGCGCCGCGCACGCCCGCGCCGCACACGCGCACGCGCGCGCTCTCGCCAAACATCTCCGCAAGCGCGCCCGCAAAGACAAGCACGACCCGCCGCCGCCGCCAGCAGTAACTGATTCTAAGAAAAAAGAGAGTACGGCAACCGCCAATTCTGTTCCGACGACGAATGCGAATTACTACGCGCCGACGACAAACACGCAGTCCTACGACCAGTCGTACACGCAGTCCTACCAGCCGACCTGGGGCTACCAGCAGCCCCCCGGGCCCTACCAACACCACCCCCACGCCTGGCCGCAGTATCCCAACTACTATTAG
Protein
MEDETSSDNQKGSENPMDTDEGPLINENSNGLSHLLTEGEFNSVDAAVDESSTSNQCFLNAVELSAGSAGDYLENNGPRFSTDSFDIGDNAVTFPNDSLNSALTEDNSIQSDSASHKAVLDTDSRQSESQHATDSNVGEINENSGNDSNFATEHVNLNVDKSEQLEIVEPCSSPVKDPVIKQDEGKEFKNQREVDDIKEQSQDSASKIGGAEETEVVSEDELPEVQKPSIKDAENVSDDELPGPKPAELPADTEVVSEDELPASKKESRKRKPDEERSNYDPGSPTSEGESSTKKQAITKNGESKPIPAEKRPSVDEKPKKKVLPELEKYWKVVNDDPTDFTGWTYLLQYVDQESDVEAAREAYDAFLSHYPYCYGYWRKFADYEKRKGSKKKCLEVLERGLKAIPLSVDLWIHYLNHIKATKTEDHAYIRSQYERAIDACGLEFRSDRLWESYIKWEAENGSALNVTNIYDRLLATPTLGYTSHFDNFQEHVMSEPVTGVVSHAELVRLRGEVRDAAGAQPSLDLPPGEDEPVDHVASDEEAQAIKERIVAARRKIHKTTGEEVTARWTFEEGIKRPYFHVKPLERCQLKNWKAYLEWEKHHGSFKRALVLHERCLIACALYEEFWMKLIKFLEERAEQDPEVIAIEREVLERACTVHHLDKPELHLHWAHFEEVQGNTLKAAEILDRIEKTCPNLVQIQYRRVNLERRRGDYEKCSQLYETYISTSKNKSLASALAIKYARFLQHVRRQPAAARAALDDAVAKDPLNPRLHLQRLDLAINTPGVTYEELDELAQSYSRQEGADTEVSAHMAWRRRELAEELGSAAHARAAHAHARALAKHLRKRARKDKHDPPPPPAVTDSKKKESTATANSVPTTNANYYAPTTNTQSYDQSYTQSYQPTWGYQQPPGPYQHHPHAWPQYPNYY
Summary
Uniprot
H9J5Z8
A0A2A4JM39
A0A3S2TD14
A0A194RL97
A0A194PG71
A0A212ENJ8
+ More
A0A1W4X7B7 A0A1W4X761 D6WSZ2 U4U7D3 A0A2J7PR87 A0A2J7PR78 A0A2J7PR71 A0A232FBM5 K7IUU3 A0A067R7B3 A0A1B6C0I2 A0A1B6EH60 E0VIE7 A0A195B657 A0A158NI42 E0VIE6 E9IBA9 A0A195EG00 F4WGD5 A0A195F0W3 A0A026WTH6 E2BV81 E2ALU2 A0A088A060 T1HUU8 A0A2P8ZK66 A0A2A3E534
A0A1W4X7B7 A0A1W4X761 D6WSZ2 U4U7D3 A0A2J7PR87 A0A2J7PR78 A0A2J7PR71 A0A232FBM5 K7IUU3 A0A067R7B3 A0A1B6C0I2 A0A1B6EH60 E0VIE7 A0A195B657 A0A158NI42 E0VIE6 E9IBA9 A0A195EG00 F4WGD5 A0A195F0W3 A0A026WTH6 E2BV81 E2ALU2 A0A088A060 T1HUU8 A0A2P8ZK66 A0A2A3E534
Pubmed
EMBL
BABH01029932
NWSH01001069
PCG72788.1
RSAL01000329
RVE42503.1
KQ460045
+ More
KPJ18194.1 KQ459604 KPI92297.1 AGBW02013634 OWR43072.1 KQ971352 EFA06677.2 KB632046 ERL88258.1 NEVH01022633 PNF18853.1 PNF18843.1 PNF18840.1 NNAY01000550 OXU27737.1 AAZX01002640 KK852653 KDR19350.1 GEDC01030290 JAS07008.1 GEDC01000057 JAS37241.1 DS235199 EEB13153.1 KQ976579 KYM79991.1 ADTU01016267 EEB13152.1 GL762111 EFZ22139.1 KQ978957 KYN27091.1 GL888128 EGI66902.1 KQ981897 KYN33749.1 KK107111 QOIP01000011 EZA58966.1 RLU16541.1 GL450824 EFN80373.1 GL440629 EFN65612.1 ACPB03018896 PYGN01000032 PSN56889.1 KZ288379 PBC26604.1
KPJ18194.1 KQ459604 KPI92297.1 AGBW02013634 OWR43072.1 KQ971352 EFA06677.2 KB632046 ERL88258.1 NEVH01022633 PNF18853.1 PNF18843.1 PNF18840.1 NNAY01000550 OXU27737.1 AAZX01002640 KK852653 KDR19350.1 GEDC01030290 JAS07008.1 GEDC01000057 JAS37241.1 DS235199 EEB13153.1 KQ976579 KYM79991.1 ADTU01016267 EEB13152.1 GL762111 EFZ22139.1 KQ978957 KYN27091.1 GL888128 EGI66902.1 KQ981897 KYN33749.1 KK107111 QOIP01000011 EZA58966.1 RLU16541.1 GL450824 EFN80373.1 GL440629 EFN65612.1 ACPB03018896 PYGN01000032 PSN56889.1 KZ288379 PBC26604.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053240
UP000053268
UP000007151
+ More
UP000192223 UP000007266 UP000030742 UP000235965 UP000215335 UP000002358 UP000027135 UP000009046 UP000078540 UP000005205 UP000078492 UP000007755 UP000078541 UP000053097 UP000279307 UP000008237 UP000000311 UP000005203 UP000015103 UP000245037 UP000242457
UP000192223 UP000007266 UP000030742 UP000235965 UP000215335 UP000002358 UP000027135 UP000009046 UP000078540 UP000005205 UP000078492 UP000007755 UP000078541 UP000053097 UP000279307 UP000008237 UP000000311 UP000005203 UP000015103 UP000245037 UP000242457
PRIDE
SUPFAM
SSF48452
SSF48452
Gene 3D
ProteinModelPortal
H9J5Z8
A0A2A4JM39
A0A3S2TD14
A0A194RL97
A0A194PG71
A0A212ENJ8
+ More
A0A1W4X7B7 A0A1W4X761 D6WSZ2 U4U7D3 A0A2J7PR87 A0A2J7PR78 A0A2J7PR71 A0A232FBM5 K7IUU3 A0A067R7B3 A0A1B6C0I2 A0A1B6EH60 E0VIE7 A0A195B657 A0A158NI42 E0VIE6 E9IBA9 A0A195EG00 F4WGD5 A0A195F0W3 A0A026WTH6 E2BV81 E2ALU2 A0A088A060 T1HUU8 A0A2P8ZK66 A0A2A3E534
A0A1W4X7B7 A0A1W4X761 D6WSZ2 U4U7D3 A0A2J7PR87 A0A2J7PR78 A0A2J7PR71 A0A232FBM5 K7IUU3 A0A067R7B3 A0A1B6C0I2 A0A1B6EH60 E0VIE7 A0A195B657 A0A158NI42 E0VIE6 E9IBA9 A0A195EG00 F4WGD5 A0A195F0W3 A0A026WTH6 E2BV81 E2ALU2 A0A088A060 T1HUU8 A0A2P8ZK66 A0A2A3E534
PDB
6G70
E-value=3.0258e-104,
Score=971
Ontologies
GO
Topology
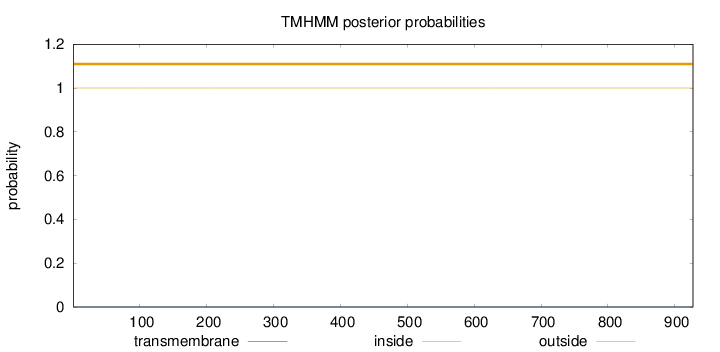
Length:
927
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00003
outside
1 - 927
Population Genetic Test Statistics
Pi
254.159982
Theta
186.587496
Tajima's D
1.397039
CLR
0.031959
CSRT
0.762161891905405
Interpretation
Uncertain
