Gene
KWMTBOMO15175 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA004977
Annotation
PREDICTED:_39S_ribosomal_protein_L9?_mitochondrial_[Plutella_xylostella]
Full name
Angiotensin-converting enzyme
+ More
39S ribosomal protein L9, mitochondrial
39S ribosomal protein L9, mitochondrial
Location in the cell
Mitochondrial Reliability : 3.174
Sequence
CDS
ATGTTTGGGCTGCGGGCTTCTGTTATGAAATCAGCTATTATTGTGCCGACAATGTTTTCACAACAAACAAGGAACACTTTTATCTTGCGACGCAGATGGCCACCACCACTTCACAAGAAAGGTGGTAAAGTGCCTAAAATGAAAGGCAGACACTTCGTTTATGACTTAGTAGAAGATACTAGTGTTAAAAAGAAACCAGACATAAGAATTGTTTTAAATCAATTTGTTGAAGGAGTTGGAACAACTGGAGATGTTTTAACACTACATTTGAACAAAGCATATGAAAACTTCATATTGCCAGGTCTTGCTGTGTATGCCAATCCAGAAAATTTGGAAAAATATAAAACCTATGAAAAACGTCCTCTAGAAGAAAATACTCACAGCTCTCCATTTGTGAAAAGGACAATGGATTGTTTACACAGATTAGTACTTCGTGTAACAATGAGCAATTCTGAACCATGGACTCTACAGCCCTGGCATCTCAAAGCTTCATTTCGCAAGTCAGGCTTCGTAGTTCCTGAGAGTGCTATTGAAATGCCTCCAGTGCAAATCAAGGGCCCAGATCCAAATTTACTCGACAAAGAATTTTATGTTACAGTAACAATAAACAAAAAGGAGAAAGTACATGTCAGATGTAGAATATATCACTGGGCAGCAGGTCAAGAGCAATTACCTTTTGAAGAAAGTTACTGGAAGAAACCTAAACAGCCACTGATACCCGAACAAGCAGAAGTACTTCAAAAATTTCCTTACCAAAATGAATAG
Protein
MFGLRASVMKSAIIVPTMFSQQTRNTFILRRRWPPPLHKKGGKVPKMKGRHFVYDLVEDTSVKKKPDIRIVLNQFVEGVGTTGDVLTLHLNKAYENFILPGLAVYANPENLEKYKTYEKRPLEENTHSSPFVKRTMDCLHRLVLRVTMSNSEPWTLQPWHLKASFRKSGFVVPESAIEMPPVQIKGPDPNLLDKEFYVTVTINKKEKVHVRCRIYHWAAGQEQLPFEESYWKKPKQPLIPEQAEVLQKFPYQNE
Summary
Cofactor
Zn(2+)
Subunit
Component of the mitochondrial ribosome large subunit (39S) which comprises a 16S rRNA and about 50 distinct proteins.
Similarity
Belongs to the peptidase M2 family.
Belongs to the bacterial ribosomal protein bL9 family.
Belongs to the bacterial ribosomal protein bL9 family.
Keywords
Complete proteome
Mitochondrion
Reference proteome
Ribonucleoprotein
Ribosomal protein
Transit peptide
Feature
chain 39S ribosomal protein L9, mitochondrial
Uniprot
H9J636
A0A1E1W5X2
A0A2A4JLX8
A0A212ENL7
A0A2H1WW27
A0A194RLK3
+ More
A0A194PFY4 A0A182XGN5 A0A182K5G3 A0A182KZX6 Q7QI21 A0A182V1Y2 A0A182IDL9 V5GQV4 A0A182TRS8 A0A182PKY9 A0A182J194 J3JXJ6 A0A1Y9HEK4 A0A182WAR3 A0A023ENK8 A0A182H2W7 N6TDB9 A0A182LRJ7 A0A182Q3R6 A0A084WF87 A0A1Q3FA39 A0A0L0CJ46 A0A182NC47 W5JBE9 B0X1W2 A0A2M3ZB56 A0A2M4APK1 Q16Q66 A0A182YS69 A0A2M4BVS9 A0A1I8MIW3 A0A336M107 A0A336KCL5 A0A0K8W459 A0A0M4EH23 A0A084VRP3 A0A0P4VQX1 A0A195BW33 A0A182F9Y7 A0A158NCT2 A0A0A1XTB0 A0A034WIH4 A0A0T6BCR3 A0A0C9PSK5 A0A1I8P9H6 W8C2F5 Q9VF89 A0A1Y1MSE5 B4MBJ8 D6WTJ6 B4HDX8 B4QYJ4 A0A1W4VSB3 B3MSN0 A0A069DQ48 A0A195F9J0 K7J419 A0A1A9YQ11 A0A0K8TRN2 A0A1B0B503 A0A154PJ61 B4N7Z1 B3P0U8 A0A0V0G983 A0A1A9UFI2 B4JS09 Q299G3 B4G550 A0A224XW01 A0A1A9ZT30 B4PS50 D3TRL6 A0A151J6B0 B4KCX8 F4X831 E9IFB0 A0A0A9YF39 A0A232FEM9 A0A146LK09 A0A151WFL0 A0A088AB54 A0A0M9A4E7 E2AG83 A0A3B0KMH1 A0A067R6A9 A0A2A3ERB6 A0A0N7ZDP8 A0A2J7QF79 A0A1J1HPD0 A0A026VXX1 A0A1L8DRF5 A0A1L8DRQ0 A0A195CEB0 A0A2P8XY89
A0A194PFY4 A0A182XGN5 A0A182K5G3 A0A182KZX6 Q7QI21 A0A182V1Y2 A0A182IDL9 V5GQV4 A0A182TRS8 A0A182PKY9 A0A182J194 J3JXJ6 A0A1Y9HEK4 A0A182WAR3 A0A023ENK8 A0A182H2W7 N6TDB9 A0A182LRJ7 A0A182Q3R6 A0A084WF87 A0A1Q3FA39 A0A0L0CJ46 A0A182NC47 W5JBE9 B0X1W2 A0A2M3ZB56 A0A2M4APK1 Q16Q66 A0A182YS69 A0A2M4BVS9 A0A1I8MIW3 A0A336M107 A0A336KCL5 A0A0K8W459 A0A0M4EH23 A0A084VRP3 A0A0P4VQX1 A0A195BW33 A0A182F9Y7 A0A158NCT2 A0A0A1XTB0 A0A034WIH4 A0A0T6BCR3 A0A0C9PSK5 A0A1I8P9H6 W8C2F5 Q9VF89 A0A1Y1MSE5 B4MBJ8 D6WTJ6 B4HDX8 B4QYJ4 A0A1W4VSB3 B3MSN0 A0A069DQ48 A0A195F9J0 K7J419 A0A1A9YQ11 A0A0K8TRN2 A0A1B0B503 A0A154PJ61 B4N7Z1 B3P0U8 A0A0V0G983 A0A1A9UFI2 B4JS09 Q299G3 B4G550 A0A224XW01 A0A1A9ZT30 B4PS50 D3TRL6 A0A151J6B0 B4KCX8 F4X831 E9IFB0 A0A0A9YF39 A0A232FEM9 A0A146LK09 A0A151WFL0 A0A088AB54 A0A0M9A4E7 E2AG83 A0A3B0KMH1 A0A067R6A9 A0A2A3ERB6 A0A0N7ZDP8 A0A2J7QF79 A0A1J1HPD0 A0A026VXX1 A0A1L8DRF5 A0A1L8DRQ0 A0A195CEB0 A0A2P8XY89
EC Number
3.4.-.-
Pubmed
19121390
22118469
26354079
20966253
12364791
14747013
+ More
17210077 22516182 24945155 26483478 23537049 24438588 26108605 20920257 23761445 17510324 25244985 25315136 27129103 21347285 25830018 25348373 24495485 10731132 12537572 12537569 11279069 28004739 17994087 18362917 19820115 26334808 20075255 26369729 15632085 17550304 20353571 21719571 21282665 25401762 28648823 26823975 20798317 24845553 24508170 30249741 29403074
17210077 22516182 24945155 26483478 23537049 24438588 26108605 20920257 23761445 17510324 25244985 25315136 27129103 21347285 25830018 25348373 24495485 10731132 12537572 12537569 11279069 28004739 17994087 18362917 19820115 26334808 20075255 26369729 15632085 17550304 20353571 21719571 21282665 25401762 28648823 26823975 20798317 24845553 24508170 30249741 29403074
EMBL
BABH01029932
GDQN01008677
JAT82377.1
NWSH01001069
PCG72786.1
AGBW02013634
+ More
OWR43073.1 ODYU01011451 SOQ57192.1 KQ460045 KPJ18195.1 KQ459604 KPI92296.1 AAAB01008811 EAA04926.5 APCN01004735 GALX01005823 JAB62643.1 BT127965 AEE62927.1 GAPW01003599 JAC09999.1 JXUM01107110 KQ565113 KXJ71322.1 APGK01042108 KB741002 KB631984 ENN75723.1 ERL87712.1 AXCM01000075 AXCN02001427 ATLV01023319 KE525342 KFB48881.1 GFDL01010656 JAV24389.1 JRES01000310 KNC32403.1 ADMH02001983 ETN60084.1 DS232272 EDS38892.1 GGFM01004980 MBW25731.1 GGFK01009405 MBW42726.1 CH477759 EAT36540.1 GGFJ01007961 MBW57102.1 UFQS01000301 UFQT01000301 SSX02637.1 SSX23011.1 SSX02634.1 SSX23008.1 GDHF01006311 JAI46003.1 CP012526 ALC47791.1 ATLV01015761 KE525036 KFB40637.1 GDKW01003058 JAI53537.1 KQ976396 KYM92844.1 ADTU01011776 GBXI01000459 JAD13833.1 GAKP01004825 JAC54127.1 LJIG01001831 KRT85109.1 GBYB01004318 JAG74085.1 GAMC01006569 JAB99986.1 AE014297 AY071462 AY118435 AAL49084.1 GEZM01024084 GEZM01024083 GEZM01024082 JAV87998.1 CH940656 EDW58469.1 KQ971354 EFA07188.1 CH480815 EDW42070.1 CM000364 EDX12839.1 CH902623 EDV30270.1 GBGD01002696 JAC86193.1 KQ981727 KYN37108.1 AAZX01000992 GDAI01000579 JAI17024.1 JXJN01008576 KQ434905 KZC11240.1 CH964232 EDW81242.1 CH954181 EDV48996.1 GECL01001484 JAP04640.1 CH916373 EDV94549.1 CM000070 EAL27740.2 CH479179 EDW24716.1 GFTR01003776 JAW12650.1 CM000160 EDW97470.1 CCAG010004727 EZ424068 ADD20344.1 KQ979863 KYN18763.1 CH933806 EDW17000.1 GL888912 EGI57380.1 GL762806 EFZ20743.1 GBHO01013876 GBRD01006881 JAG29728.1 JAG58940.1 NNAY01000320 OXU29244.1 GDHC01011014 JAQ07615.1 KQ983211 KYQ46632.1 KQ435736 KOX77007.1 GL439266 EFN67574.1 OUUW01000013 SPP87779.1 KK852670 KDR18805.1 KZ288194 PBC33812.1 GDRN01015265 JAI67848.1 NEVH01015301 PNF27234.1 CVRI01000006 CRK88334.1 KK107606 QOIP01000007 EZA48530.1 RLU20349.1 GFDF01005038 JAV09046.1 GFDF01005054 JAV09030.1 KQ977873 KYM99137.1 PYGN01001171 PSN36982.1
OWR43073.1 ODYU01011451 SOQ57192.1 KQ460045 KPJ18195.1 KQ459604 KPI92296.1 AAAB01008811 EAA04926.5 APCN01004735 GALX01005823 JAB62643.1 BT127965 AEE62927.1 GAPW01003599 JAC09999.1 JXUM01107110 KQ565113 KXJ71322.1 APGK01042108 KB741002 KB631984 ENN75723.1 ERL87712.1 AXCM01000075 AXCN02001427 ATLV01023319 KE525342 KFB48881.1 GFDL01010656 JAV24389.1 JRES01000310 KNC32403.1 ADMH02001983 ETN60084.1 DS232272 EDS38892.1 GGFM01004980 MBW25731.1 GGFK01009405 MBW42726.1 CH477759 EAT36540.1 GGFJ01007961 MBW57102.1 UFQS01000301 UFQT01000301 SSX02637.1 SSX23011.1 SSX02634.1 SSX23008.1 GDHF01006311 JAI46003.1 CP012526 ALC47791.1 ATLV01015761 KE525036 KFB40637.1 GDKW01003058 JAI53537.1 KQ976396 KYM92844.1 ADTU01011776 GBXI01000459 JAD13833.1 GAKP01004825 JAC54127.1 LJIG01001831 KRT85109.1 GBYB01004318 JAG74085.1 GAMC01006569 JAB99986.1 AE014297 AY071462 AY118435 AAL49084.1 GEZM01024084 GEZM01024083 GEZM01024082 JAV87998.1 CH940656 EDW58469.1 KQ971354 EFA07188.1 CH480815 EDW42070.1 CM000364 EDX12839.1 CH902623 EDV30270.1 GBGD01002696 JAC86193.1 KQ981727 KYN37108.1 AAZX01000992 GDAI01000579 JAI17024.1 JXJN01008576 KQ434905 KZC11240.1 CH964232 EDW81242.1 CH954181 EDV48996.1 GECL01001484 JAP04640.1 CH916373 EDV94549.1 CM000070 EAL27740.2 CH479179 EDW24716.1 GFTR01003776 JAW12650.1 CM000160 EDW97470.1 CCAG010004727 EZ424068 ADD20344.1 KQ979863 KYN18763.1 CH933806 EDW17000.1 GL888912 EGI57380.1 GL762806 EFZ20743.1 GBHO01013876 GBRD01006881 JAG29728.1 JAG58940.1 NNAY01000320 OXU29244.1 GDHC01011014 JAQ07615.1 KQ983211 KYQ46632.1 KQ435736 KOX77007.1 GL439266 EFN67574.1 OUUW01000013 SPP87779.1 KK852670 KDR18805.1 KZ288194 PBC33812.1 GDRN01015265 JAI67848.1 NEVH01015301 PNF27234.1 CVRI01000006 CRK88334.1 KK107606 QOIP01000007 EZA48530.1 RLU20349.1 GFDF01005038 JAV09046.1 GFDF01005054 JAV09030.1 KQ977873 KYM99137.1 PYGN01001171 PSN36982.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000053268
UP000076407
+ More
UP000075881 UP000075882 UP000007062 UP000075903 UP000075840 UP000075902 UP000075885 UP000075880 UP000075900 UP000075920 UP000069940 UP000249989 UP000019118 UP000030742 UP000075883 UP000075886 UP000030765 UP000037069 UP000075884 UP000000673 UP000002320 UP000008820 UP000076408 UP000095301 UP000092553 UP000078540 UP000069272 UP000005205 UP000095300 UP000000803 UP000008792 UP000007266 UP000001292 UP000000304 UP000192221 UP000007801 UP000078541 UP000002358 UP000092443 UP000092460 UP000076502 UP000007798 UP000008711 UP000078200 UP000001070 UP000001819 UP000008744 UP000092445 UP000002282 UP000092444 UP000078492 UP000009192 UP000007755 UP000215335 UP000075809 UP000005203 UP000053105 UP000000311 UP000268350 UP000027135 UP000242457 UP000235965 UP000183832 UP000053097 UP000279307 UP000078542 UP000245037
UP000075881 UP000075882 UP000007062 UP000075903 UP000075840 UP000075902 UP000075885 UP000075880 UP000075900 UP000075920 UP000069940 UP000249989 UP000019118 UP000030742 UP000075883 UP000075886 UP000030765 UP000037069 UP000075884 UP000000673 UP000002320 UP000008820 UP000076408 UP000095301 UP000092553 UP000078540 UP000069272 UP000005205 UP000095300 UP000000803 UP000008792 UP000007266 UP000001292 UP000000304 UP000192221 UP000007801 UP000078541 UP000002358 UP000092443 UP000092460 UP000076502 UP000007798 UP000008711 UP000078200 UP000001070 UP000001819 UP000008744 UP000092445 UP000002282 UP000092444 UP000078492 UP000009192 UP000007755 UP000215335 UP000075809 UP000005203 UP000053105 UP000000311 UP000268350 UP000027135 UP000242457 UP000235965 UP000183832 UP000053097 UP000279307 UP000078542 UP000245037
Interpro
SUPFAM
SSF55658
SSF55658
Gene 3D
CDD
ProteinModelPortal
H9J636
A0A1E1W5X2
A0A2A4JLX8
A0A212ENL7
A0A2H1WW27
A0A194RLK3
+ More
A0A194PFY4 A0A182XGN5 A0A182K5G3 A0A182KZX6 Q7QI21 A0A182V1Y2 A0A182IDL9 V5GQV4 A0A182TRS8 A0A182PKY9 A0A182J194 J3JXJ6 A0A1Y9HEK4 A0A182WAR3 A0A023ENK8 A0A182H2W7 N6TDB9 A0A182LRJ7 A0A182Q3R6 A0A084WF87 A0A1Q3FA39 A0A0L0CJ46 A0A182NC47 W5JBE9 B0X1W2 A0A2M3ZB56 A0A2M4APK1 Q16Q66 A0A182YS69 A0A2M4BVS9 A0A1I8MIW3 A0A336M107 A0A336KCL5 A0A0K8W459 A0A0M4EH23 A0A084VRP3 A0A0P4VQX1 A0A195BW33 A0A182F9Y7 A0A158NCT2 A0A0A1XTB0 A0A034WIH4 A0A0T6BCR3 A0A0C9PSK5 A0A1I8P9H6 W8C2F5 Q9VF89 A0A1Y1MSE5 B4MBJ8 D6WTJ6 B4HDX8 B4QYJ4 A0A1W4VSB3 B3MSN0 A0A069DQ48 A0A195F9J0 K7J419 A0A1A9YQ11 A0A0K8TRN2 A0A1B0B503 A0A154PJ61 B4N7Z1 B3P0U8 A0A0V0G983 A0A1A9UFI2 B4JS09 Q299G3 B4G550 A0A224XW01 A0A1A9ZT30 B4PS50 D3TRL6 A0A151J6B0 B4KCX8 F4X831 E9IFB0 A0A0A9YF39 A0A232FEM9 A0A146LK09 A0A151WFL0 A0A088AB54 A0A0M9A4E7 E2AG83 A0A3B0KMH1 A0A067R6A9 A0A2A3ERB6 A0A0N7ZDP8 A0A2J7QF79 A0A1J1HPD0 A0A026VXX1 A0A1L8DRF5 A0A1L8DRQ0 A0A195CEB0 A0A2P8XY89
A0A194PFY4 A0A182XGN5 A0A182K5G3 A0A182KZX6 Q7QI21 A0A182V1Y2 A0A182IDL9 V5GQV4 A0A182TRS8 A0A182PKY9 A0A182J194 J3JXJ6 A0A1Y9HEK4 A0A182WAR3 A0A023ENK8 A0A182H2W7 N6TDB9 A0A182LRJ7 A0A182Q3R6 A0A084WF87 A0A1Q3FA39 A0A0L0CJ46 A0A182NC47 W5JBE9 B0X1W2 A0A2M3ZB56 A0A2M4APK1 Q16Q66 A0A182YS69 A0A2M4BVS9 A0A1I8MIW3 A0A336M107 A0A336KCL5 A0A0K8W459 A0A0M4EH23 A0A084VRP3 A0A0P4VQX1 A0A195BW33 A0A182F9Y7 A0A158NCT2 A0A0A1XTB0 A0A034WIH4 A0A0T6BCR3 A0A0C9PSK5 A0A1I8P9H6 W8C2F5 Q9VF89 A0A1Y1MSE5 B4MBJ8 D6WTJ6 B4HDX8 B4QYJ4 A0A1W4VSB3 B3MSN0 A0A069DQ48 A0A195F9J0 K7J419 A0A1A9YQ11 A0A0K8TRN2 A0A1B0B503 A0A154PJ61 B4N7Z1 B3P0U8 A0A0V0G983 A0A1A9UFI2 B4JS09 Q299G3 B4G550 A0A224XW01 A0A1A9ZT30 B4PS50 D3TRL6 A0A151J6B0 B4KCX8 F4X831 E9IFB0 A0A0A9YF39 A0A232FEM9 A0A146LK09 A0A151WFL0 A0A088AB54 A0A0M9A4E7 E2AG83 A0A3B0KMH1 A0A067R6A9 A0A2A3ERB6 A0A0N7ZDP8 A0A2J7QF79 A0A1J1HPD0 A0A026VXX1 A0A1L8DRF5 A0A1L8DRQ0 A0A195CEB0 A0A2P8XY89
PDB
6GB2
E-value=6.24141e-16,
Score=203
Ontologies
GO
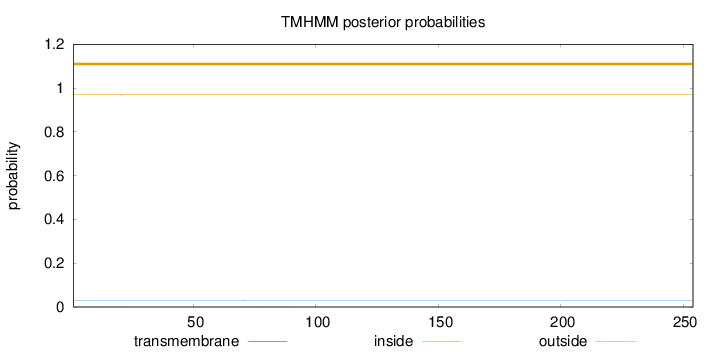
Topology
Subcellular location
Mitochondrion
Length:
254
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01616
Exp number, first 60 AAs:
0.00986
Total prob of N-in:
0.03114
outside
1 - 254
Population Genetic Test Statistics
Pi
244.152033
Theta
203.060889
Tajima's D
0.621472
CLR
0.04163
CSRT
0.548372581370931
Interpretation
Uncertain
