Pre Gene Modal
BGIBMGA004982
Annotation
PREDICTED:_polyglutamine-binding_protein_1_[Papilio_machaon]
Location in the cell
Nuclear Reliability : 3.764
Sequence
CDS
ATGCCTTTACCGCCGGCGTTAGCTGCTCGTTTAGCTAAAAGAGGTCTTTTGAAACAAAATGCAGATTCCACCGCTGTCGCATCTGTTGGTCAAATCAACGAAGAAATAATTGCAGAAGATTATGATAGTAAGCCTAACGAAAATGGAAAAGATGAGCCTTGGGAAGGCTTTGTCGATCCTGTCAAAGGCCATCGAGGATGCCCTAATAAAAGTAATATTTATCATGAATGTTCTACTTTTTGTATTAAAAGATGGAAGCAAGGTAAACTGGTACCGACTGAGACATATTTAGAACACAAGGAGAAAGTTTTAGAGCTATGGCCCTTGCCACCAGGATGGGAAGAAGTGTTTGATGAAGGTTATGGTCAACATTATTTTTGGAATGTGCACACTAATCTGGTTTCGTGGATACCACCCGGACATCCCCGTGCTGTGCCTACGGAATCTGCCGCACAATTGCGCGAAGAACGTCTGATGAAAGAAGGCGACGAAAGTGATGAAAGCAGTGAGGCCTCGGATCAGGAAGTTCCGCAGATTCAAAAAGATAAGAAACCTGATCGCCGAGAAAGAGAAAAGGAACATTCACATCATAGAGACAAAAAAAGGCTCAGGGTCAAAGATAATGACTTGGATCCAATGGACCCAGCATCTTACTCTGATGTACCTAGAGGTAATTGGACATCAGGTTTAGATACACATTCTAAAACAGGTGTAGATACCACAGCCTCTGGTTCACTCTATCAAATGAGGCCCTATCCAGCACCTGGAGCAATACTTGCAGCTAATGCAAAACAACATTCTCCACCACCTTCACCATAA
Protein
MPLPPALAARLAKRGLLKQNADSTAVASVGQINEEIIAEDYDSKPNENGKDEPWEGFVDPVKGHRGCPNKSNIYHECSTFCIKRWKQGKLVPTETYLEHKEKVLELWPLPPGWEEVFDEGYGQHYFWNVHTNLVSWIPPGHPRAVPTESAAQLREERLMKEGDESDESSEASDQEVPQIQKDKKPDRREREKEHSHHRDKKRLRVKDNDLDPMDPASYSDVPRGNWTSGLDTHSKTGVDTTASGSLYQMRPYPAPGAILAANAKQHSPPPSP
Summary
Uniprot
H9J641
A0A194RLA8
A0A1E1W0U1
A0A194PHU1
A0A2H1WRA7
A0A2A4JMI1
+ More
A0A0L7KND3 A0A3S2NFY7 S4P4D2 A0A212F1M9 A0A1S3DHU9 A0A224XTZ9 A0A0P4VQS9 R4FM17 A0A2R7WIF2 E0W331 J9K136 C4WSG7 A0A0C9PUG0 A0A0A9YR75 A0A2H8TD72 A0A069DRZ6 A0A0N0BCN9 A0A026WPK8 A0A151IEH6 A0A151X3Q2 A0A087ZQX7 A0A195EEX7 A0A2A3EKU4 A0A0L7RC77 A0A195BQ27 A0A195EST0 A0A158NTJ7 F4WQW2 E2C4F1 A0A0J7KWI9 A0A1B0GJ48 A0A1B6CRD3 A0A1L8DG11 A0A1B6HJX7 A0A232EUI5 K7J773 E9FZ08 A0A1B6EUH7 A0A2J7RTD2 A0A1B6KDX2 A0A1B6KEL8 A0A164Y8U1 A0A067QN72 A0A0P5IT66 A0A0P5NWL0 A0A293MZ93 A0A0P5Q252 E2AFX7 A0A0P5FJB5 A0A0P5QR97 A0A0P5K1G5 A0A1Z5L691 T1IT40 A0A2R5LJ02 G3MFB8 A0A1E1X9N0 A0A023GKM2 A0A131XF70 A0A0P4WUB9 B7QKP7 V5H8Y7 A0A224Z627 L7M858 A0A1E1XU70 A0A023FKN7 A0A154PLC4 A0A087U604 A0A0P5XRL5 D6WM89 A0A3P8F4Z3 A0A0P6BKH2 A0A2G9UJU2 A0A1I7XSD6 A0A226EN44 A0A1Y1K1I4 A0A1D1UWR2 T1FNT2 A0A3S3P4V3 T1L017 A0A1B0GQN4 A0A336K7P6 A0A016VS23 A0A158R2K2 A0A0R3PQD9 J3JUS8 A0A0K0DMY9 A0A310S937 A0A0C2CAY4 A0A0C9S1J8 W2T5M9 K1R639 A0A0D8XLD1 A0A0V1N9E7 A0A1Y3EIC8
A0A0L7KND3 A0A3S2NFY7 S4P4D2 A0A212F1M9 A0A1S3DHU9 A0A224XTZ9 A0A0P4VQS9 R4FM17 A0A2R7WIF2 E0W331 J9K136 C4WSG7 A0A0C9PUG0 A0A0A9YR75 A0A2H8TD72 A0A069DRZ6 A0A0N0BCN9 A0A026WPK8 A0A151IEH6 A0A151X3Q2 A0A087ZQX7 A0A195EEX7 A0A2A3EKU4 A0A0L7RC77 A0A195BQ27 A0A195EST0 A0A158NTJ7 F4WQW2 E2C4F1 A0A0J7KWI9 A0A1B0GJ48 A0A1B6CRD3 A0A1L8DG11 A0A1B6HJX7 A0A232EUI5 K7J773 E9FZ08 A0A1B6EUH7 A0A2J7RTD2 A0A1B6KDX2 A0A1B6KEL8 A0A164Y8U1 A0A067QN72 A0A0P5IT66 A0A0P5NWL0 A0A293MZ93 A0A0P5Q252 E2AFX7 A0A0P5FJB5 A0A0P5QR97 A0A0P5K1G5 A0A1Z5L691 T1IT40 A0A2R5LJ02 G3MFB8 A0A1E1X9N0 A0A023GKM2 A0A131XF70 A0A0P4WUB9 B7QKP7 V5H8Y7 A0A224Z627 L7M858 A0A1E1XU70 A0A023FKN7 A0A154PLC4 A0A087U604 A0A0P5XRL5 D6WM89 A0A3P8F4Z3 A0A0P6BKH2 A0A2G9UJU2 A0A1I7XSD6 A0A226EN44 A0A1Y1K1I4 A0A1D1UWR2 T1FNT2 A0A3S3P4V3 T1L017 A0A1B0GQN4 A0A336K7P6 A0A016VS23 A0A158R2K2 A0A0R3PQD9 J3JUS8 A0A0K0DMY9 A0A310S937 A0A0C2CAY4 A0A0C9S1J8 W2T5M9 K1R639 A0A0D8XLD1 A0A0V1N9E7 A0A1Y3EIC8
Pubmed
19121390
26354079
26227816
23622113
22118469
27129103
+ More
20566863 25401762 26823975 26334808 24508170 30249741 21347285 21719571 20798317 28648823 20075255 21292972 24845553 28528879 22216098 28503490 28049606 25765539 28797301 25576852 29209593 18362917 19820115 23874975 28004739 27649274 23254933 25730766 22516182 26131772 22992520 26856411
20566863 25401762 26823975 26334808 24508170 30249741 21347285 21719571 20798317 28648823 20075255 21292972 24845553 28528879 22216098 28503490 28049606 25765539 28797301 25576852 29209593 18362917 19820115 23874975 28004739 27649274 23254933 25730766 22516182 26131772 22992520 26856411
EMBL
BABH01029940
KQ460045
KPJ18204.1
GDQN01010449
JAT80605.1
KQ459604
+ More
KPI92289.1 ODYU01010411 SOQ55492.1 NWSH01001069 PCG72794.1 JTDY01008322 KOB64625.1 RSAL01000053 RVE50098.1 GAIX01006024 JAA86536.1 AGBW02010844 OWR47642.1 GFTR01004434 JAW11992.1 GDKW01001802 JAI54793.1 ACPB03006429 GAHY01001899 JAA75611.1 KK854722 PTY18175.1 DS235882 EEB20037.1 ABLF02017871 ABLF02017872 AK340182 BAH70837.1 GBYB01005013 JAG74780.1 GBHO01008047 GBRD01009900 GDHC01015924 JAG35557.1 JAG55924.1 JAQ02705.1 GFXV01000248 MBW12053.1 GBGD01002492 JAC86397.1 KQ435911 KOX68914.1 KK107152 QOIP01000007 EZA57024.1 RLU20723.1 KQ977870 KYM99230.1 KQ982562 KYQ54890.1 KQ978983 KYN26793.1 KZ288226 PBC31882.1 KQ414616 KOC68433.1 KQ976424 KYM88627.1 KQ981993 KYN30939.1 ADTU01025983 GL888275 EGI63475.1 GL452462 EFN77188.1 LBMM01002588 KMQ94609.1 AJWK01019308 GEDC01021172 GEDC01011979 JAS16126.1 JAS25319.1 GFDF01008692 JAV05392.1 GECU01032744 GECU01024481 JAS74962.1 JAS83225.1 NNAY01002129 OXU22001.1 GL732527 EFX87679.1 GECZ01028209 JAS41560.1 NEVH01000006 PNF44084.1 GEBQ01030382 JAT09595.1 GEBQ01030065 JAT09912.1 LRGB01000930 KZS14997.1 KK853217 KDR09757.1 GDIQ01209418 JAK42307.1 GDIQ01143792 JAL07934.1 GFWV01020523 MAA45251.1 GDIQ01126805 JAL24921.1 GL439180 EFN67653.1 GDIQ01256228 JAJ95496.1 GDIQ01120456 JAL31270.1 GDIQ01191118 JAK60607.1 GFJQ02004044 JAW02926.1 AFFK01019071 GGLE01005292 MBY09418.1 JO840569 AEO32186.1 GFAC01003223 JAT95965.1 GBBM01002068 JAC33350.1 GEFH01003831 JAP64750.1 GDIP01253029 JAI70372.1 ABJB010231419 ABJB010741897 DS961460 EEC19419.1 GANP01010869 JAB73599.1 GFPF01010688 MAA21834.1 GACK01005686 JAA59348.1 GFAA01000612 JAU02823.1 GBBK01003254 JAC21228.1 KQ434960 KZC12633.1 KK118353 KFM72793.1 GDIP01068318 JAM35397.1 KQ971343 EFA04236.1 UZAH01032019 VDP19169.1 GDIP01021109 JAM82606.1 KZ346246 PIO70515.1 LNIX01000002 OXA59083.1 GEZM01095841 JAV55303.1 BDGG01000002 GAU93030.1 AMQM01008205 KB097736 ESN90976.1 NCKU01010194 RWS00929.1 CAEY01000760 AJVK01007422 UFQS01000107 UFQT01000107 SSW99678.1 SSX20058.1 KE124926 JARK01001341 EPB74842.1 EYC30096.1 UYSL01022073 VDL79764.1 UYYA01004051 VDM59140.1 BT126993 AEE61955.1 KQ769179 OAD52941.1 KN768185 KIH47012.1 GBZX01002026 JAG90714.1 KI660200 ETN76914.1 JH816011 EKC41228.1 KN716408 KJH45438.1 JYDO01000002 KRZ80453.1 LVZM01015336 OUC43329.1
KPI92289.1 ODYU01010411 SOQ55492.1 NWSH01001069 PCG72794.1 JTDY01008322 KOB64625.1 RSAL01000053 RVE50098.1 GAIX01006024 JAA86536.1 AGBW02010844 OWR47642.1 GFTR01004434 JAW11992.1 GDKW01001802 JAI54793.1 ACPB03006429 GAHY01001899 JAA75611.1 KK854722 PTY18175.1 DS235882 EEB20037.1 ABLF02017871 ABLF02017872 AK340182 BAH70837.1 GBYB01005013 JAG74780.1 GBHO01008047 GBRD01009900 GDHC01015924 JAG35557.1 JAG55924.1 JAQ02705.1 GFXV01000248 MBW12053.1 GBGD01002492 JAC86397.1 KQ435911 KOX68914.1 KK107152 QOIP01000007 EZA57024.1 RLU20723.1 KQ977870 KYM99230.1 KQ982562 KYQ54890.1 KQ978983 KYN26793.1 KZ288226 PBC31882.1 KQ414616 KOC68433.1 KQ976424 KYM88627.1 KQ981993 KYN30939.1 ADTU01025983 GL888275 EGI63475.1 GL452462 EFN77188.1 LBMM01002588 KMQ94609.1 AJWK01019308 GEDC01021172 GEDC01011979 JAS16126.1 JAS25319.1 GFDF01008692 JAV05392.1 GECU01032744 GECU01024481 JAS74962.1 JAS83225.1 NNAY01002129 OXU22001.1 GL732527 EFX87679.1 GECZ01028209 JAS41560.1 NEVH01000006 PNF44084.1 GEBQ01030382 JAT09595.1 GEBQ01030065 JAT09912.1 LRGB01000930 KZS14997.1 KK853217 KDR09757.1 GDIQ01209418 JAK42307.1 GDIQ01143792 JAL07934.1 GFWV01020523 MAA45251.1 GDIQ01126805 JAL24921.1 GL439180 EFN67653.1 GDIQ01256228 JAJ95496.1 GDIQ01120456 JAL31270.1 GDIQ01191118 JAK60607.1 GFJQ02004044 JAW02926.1 AFFK01019071 GGLE01005292 MBY09418.1 JO840569 AEO32186.1 GFAC01003223 JAT95965.1 GBBM01002068 JAC33350.1 GEFH01003831 JAP64750.1 GDIP01253029 JAI70372.1 ABJB010231419 ABJB010741897 DS961460 EEC19419.1 GANP01010869 JAB73599.1 GFPF01010688 MAA21834.1 GACK01005686 JAA59348.1 GFAA01000612 JAU02823.1 GBBK01003254 JAC21228.1 KQ434960 KZC12633.1 KK118353 KFM72793.1 GDIP01068318 JAM35397.1 KQ971343 EFA04236.1 UZAH01032019 VDP19169.1 GDIP01021109 JAM82606.1 KZ346246 PIO70515.1 LNIX01000002 OXA59083.1 GEZM01095841 JAV55303.1 BDGG01000002 GAU93030.1 AMQM01008205 KB097736 ESN90976.1 NCKU01010194 RWS00929.1 CAEY01000760 AJVK01007422 UFQS01000107 UFQT01000107 SSW99678.1 SSX20058.1 KE124926 JARK01001341 EPB74842.1 EYC30096.1 UYSL01022073 VDL79764.1 UYYA01004051 VDM59140.1 BT126993 AEE61955.1 KQ769179 OAD52941.1 KN768185 KIH47012.1 GBZX01002026 JAG90714.1 KI660200 ETN76914.1 JH816011 EKC41228.1 KN716408 KJH45438.1 JYDO01000002 KRZ80453.1 LVZM01015336 OUC43329.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000218220
UP000037510
UP000283053
+ More
UP000007151 UP000079169 UP000015103 UP000009046 UP000007819 UP000053105 UP000053097 UP000279307 UP000078542 UP000075809 UP000005203 UP000078492 UP000242457 UP000053825 UP000078540 UP000078541 UP000005205 UP000007755 UP000008237 UP000036403 UP000092461 UP000215335 UP000002358 UP000000305 UP000235965 UP000076858 UP000027135 UP000000311 UP000001555 UP000076502 UP000054359 UP000007266 UP000095283 UP000198287 UP000186922 UP000015101 UP000285301 UP000015104 UP000092462 UP000024635 UP000038043 UP000271162 UP000050601 UP000267027 UP000035642 UP000005408 UP000053766 UP000054843 UP000243006
UP000007151 UP000079169 UP000015103 UP000009046 UP000007819 UP000053105 UP000053097 UP000279307 UP000078542 UP000075809 UP000005203 UP000078492 UP000242457 UP000053825 UP000078540 UP000078541 UP000005205 UP000007755 UP000008237 UP000036403 UP000092461 UP000215335 UP000002358 UP000000305 UP000235965 UP000076858 UP000027135 UP000000311 UP000001555 UP000076502 UP000054359 UP000007266 UP000095283 UP000198287 UP000186922 UP000015101 UP000285301 UP000015104 UP000092462 UP000024635 UP000038043 UP000271162 UP000050601 UP000267027 UP000035642 UP000005408 UP000053766 UP000054843 UP000243006
PRIDE
Pfam
PF00397 WW
SUPFAM
SSF51045
SSF51045
CDD
ProteinModelPortal
H9J641
A0A194RLA8
A0A1E1W0U1
A0A194PHU1
A0A2H1WRA7
A0A2A4JMI1
+ More
A0A0L7KND3 A0A3S2NFY7 S4P4D2 A0A212F1M9 A0A1S3DHU9 A0A224XTZ9 A0A0P4VQS9 R4FM17 A0A2R7WIF2 E0W331 J9K136 C4WSG7 A0A0C9PUG0 A0A0A9YR75 A0A2H8TD72 A0A069DRZ6 A0A0N0BCN9 A0A026WPK8 A0A151IEH6 A0A151X3Q2 A0A087ZQX7 A0A195EEX7 A0A2A3EKU4 A0A0L7RC77 A0A195BQ27 A0A195EST0 A0A158NTJ7 F4WQW2 E2C4F1 A0A0J7KWI9 A0A1B0GJ48 A0A1B6CRD3 A0A1L8DG11 A0A1B6HJX7 A0A232EUI5 K7J773 E9FZ08 A0A1B6EUH7 A0A2J7RTD2 A0A1B6KDX2 A0A1B6KEL8 A0A164Y8U1 A0A067QN72 A0A0P5IT66 A0A0P5NWL0 A0A293MZ93 A0A0P5Q252 E2AFX7 A0A0P5FJB5 A0A0P5QR97 A0A0P5K1G5 A0A1Z5L691 T1IT40 A0A2R5LJ02 G3MFB8 A0A1E1X9N0 A0A023GKM2 A0A131XF70 A0A0P4WUB9 B7QKP7 V5H8Y7 A0A224Z627 L7M858 A0A1E1XU70 A0A023FKN7 A0A154PLC4 A0A087U604 A0A0P5XRL5 D6WM89 A0A3P8F4Z3 A0A0P6BKH2 A0A2G9UJU2 A0A1I7XSD6 A0A226EN44 A0A1Y1K1I4 A0A1D1UWR2 T1FNT2 A0A3S3P4V3 T1L017 A0A1B0GQN4 A0A336K7P6 A0A016VS23 A0A158R2K2 A0A0R3PQD9 J3JUS8 A0A0K0DMY9 A0A310S937 A0A0C2CAY4 A0A0C9S1J8 W2T5M9 K1R639 A0A0D8XLD1 A0A0V1N9E7 A0A1Y3EIC8
A0A0L7KND3 A0A3S2NFY7 S4P4D2 A0A212F1M9 A0A1S3DHU9 A0A224XTZ9 A0A0P4VQS9 R4FM17 A0A2R7WIF2 E0W331 J9K136 C4WSG7 A0A0C9PUG0 A0A0A9YR75 A0A2H8TD72 A0A069DRZ6 A0A0N0BCN9 A0A026WPK8 A0A151IEH6 A0A151X3Q2 A0A087ZQX7 A0A195EEX7 A0A2A3EKU4 A0A0L7RC77 A0A195BQ27 A0A195EST0 A0A158NTJ7 F4WQW2 E2C4F1 A0A0J7KWI9 A0A1B0GJ48 A0A1B6CRD3 A0A1L8DG11 A0A1B6HJX7 A0A232EUI5 K7J773 E9FZ08 A0A1B6EUH7 A0A2J7RTD2 A0A1B6KDX2 A0A1B6KEL8 A0A164Y8U1 A0A067QN72 A0A0P5IT66 A0A0P5NWL0 A0A293MZ93 A0A0P5Q252 E2AFX7 A0A0P5FJB5 A0A0P5QR97 A0A0P5K1G5 A0A1Z5L691 T1IT40 A0A2R5LJ02 G3MFB8 A0A1E1X9N0 A0A023GKM2 A0A131XF70 A0A0P4WUB9 B7QKP7 V5H8Y7 A0A224Z627 L7M858 A0A1E1XU70 A0A023FKN7 A0A154PLC4 A0A087U604 A0A0P5XRL5 D6WM89 A0A3P8F4Z3 A0A0P6BKH2 A0A2G9UJU2 A0A1I7XSD6 A0A226EN44 A0A1Y1K1I4 A0A1D1UWR2 T1FNT2 A0A3S3P4V3 T1L017 A0A1B0GQN4 A0A336K7P6 A0A016VS23 A0A158R2K2 A0A0R3PQD9 J3JUS8 A0A0K0DMY9 A0A310S937 A0A0C2CAY4 A0A0C9S1J8 W2T5M9 K1R639 A0A0D8XLD1 A0A0V1N9E7 A0A1Y3EIC8
PDB
4CDO
E-value=2.50425e-06,
Score=121
Ontologies
GO
Topology
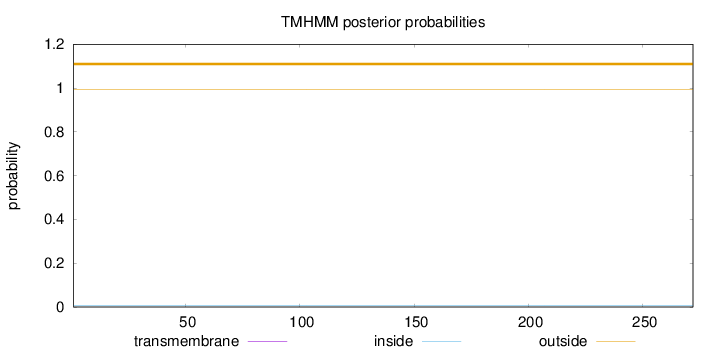
Length:
272
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00558
outside
1 - 272
Population Genetic Test Statistics
Pi
294.368486
Theta
215.301317
Tajima's D
1.156117
CLR
3.411561
CSRT
0.702114894255287
Interpretation
Uncertain
