Gene
KWMTBOMO15164
Pre Gene Modal
BGIBMGA004934
Annotation
PREDICTED:_acetylcholinesterase-like_[Papilio_machaon]
Full name
Carboxylic ester hydrolase
Location in the cell
Extracellular Reliability : 1.014
Sequence
CDS
ATGCTGGGTATTACGCACCCAGGAGCCTGTCACGGCGACGAAATGGGATACCTGTTTTATTTCTCGAGAATAAATTACAGATTAGACGAAGGTTCCACGGAATGGGCCGTATCGAAGAAAATGGTACAAATGTGGACTAACTTTGCTAAAACCGGAAATCCGACACCACCGGTCGATTACGAGTCTATTGTAGATTTTAAATGGTTACCTGTAAATGAGAGTTCGCACGTCAACTATCTAGATATCACTGGGAACTTTACATCGAAAGTGGACCCCGAGCAGAGGAGAATCAATTTTTGGGACTGGCTCTACGAGAATTTTGCGAAAAAACCTTGA
Protein
MLGITHPGACHGDEMGYLFYFSRINYRLDEGSTEWAVSKKMVQMWTNFAKTGNPTPPVDYESIVDFKWLPVNESSHVNYLDITGNFTSKVDPEQRRINFWDWLYENFAKKP
Summary
Similarity
Belongs to the type-B carboxylesterase/lipase family.
Feature
chain Carboxylic ester hydrolase
Uniprot
H9J5Z3
A0A194PFX4
A0A194RQA4
A0A212EZ22
A0A2A4J4W9
A0A1L8D6Z3
+ More
A0A2H1WR02 A0A1U9X1X2 A0A0L7LCT1 A0A0K8URL8 A0A034VR46 A0A0A1XFZ8 A0A068F517 W8B3D5 A0A182F8C8 A0A0L0CLK8 W8BIG0 A0A1I8PWQ5 A0A1B0G3F2 A0A1A9UCU4 A0A1I8PWP1 A0A1I8PWQ3 A0A1I8PWQ1 A0A1A9X418 A0A1I8PWR0 A0A1I8PWN8 B4JRH5 A0A3B0K0B9 A0A1A9ZHC1 B3LY87 A0A0M5J219 Q29BQ2 I5AN04 A0A1A9Y8U9 A0A182VX55 A0A182R3N4 A0A0R3NF95 B4GPA7 A0A0R3NF77 A0A0R1E4Y1 D3DMF0 B4PN92 A0A0R1E9H0 A0A1B0F076 A0A1W4URC9 A0A1W4URD4 B4HFG1 B3P7B4 A0A1W4V4P9 A0A182QEU4 Q8MQQ0 A0A182MJA2 Q86P08 T1PLR5 Q9VCL6 Q8IMY6 A0A0B4LHM3 A0A1I8NHY5 B4NJK8 B4M0S0 A0A0Q9WFF2 A0A0Q9WRN5 A0A1Q3G476 A0A1Q3G3Z0 A0A1Q3G408 A0A182IRH2 A0A1Q3G467 A0A1J1J596 B4KA74 A0A182VPB0 A0A0Q9XCP6 Q7PQZ9 A0A182XI96 A0A182HT83 A0A182PA59 A0A182LFB8 A0A182NA84 Q171Y4 A0A336MIB4 A0A336MV82 A0A336MQG4 A0A182YKI4 U4TU65 N6UP99 U4U2T7 B4R207 A0A1Y1LJP8 A0A2J7RMT5 A0A2J7RMR6 A0A2J7RMR7 A0A2J7RMR9 A0A1Y1MHK6 A0A1Y1MG78 A0A139WFP4 A0A2H1WPZ4 A0A0C5KC89 A0A1Y1LQE6 A0A1S4F9W3 W8EA74
A0A2H1WR02 A0A1U9X1X2 A0A0L7LCT1 A0A0K8URL8 A0A034VR46 A0A0A1XFZ8 A0A068F517 W8B3D5 A0A182F8C8 A0A0L0CLK8 W8BIG0 A0A1I8PWQ5 A0A1B0G3F2 A0A1A9UCU4 A0A1I8PWP1 A0A1I8PWQ3 A0A1I8PWQ1 A0A1A9X418 A0A1I8PWR0 A0A1I8PWN8 B4JRH5 A0A3B0K0B9 A0A1A9ZHC1 B3LY87 A0A0M5J219 Q29BQ2 I5AN04 A0A1A9Y8U9 A0A182VX55 A0A182R3N4 A0A0R3NF95 B4GPA7 A0A0R3NF77 A0A0R1E4Y1 D3DMF0 B4PN92 A0A0R1E9H0 A0A1B0F076 A0A1W4URC9 A0A1W4URD4 B4HFG1 B3P7B4 A0A1W4V4P9 A0A182QEU4 Q8MQQ0 A0A182MJA2 Q86P08 T1PLR5 Q9VCL6 Q8IMY6 A0A0B4LHM3 A0A1I8NHY5 B4NJK8 B4M0S0 A0A0Q9WFF2 A0A0Q9WRN5 A0A1Q3G476 A0A1Q3G3Z0 A0A1Q3G408 A0A182IRH2 A0A1Q3G467 A0A1J1J596 B4KA74 A0A182VPB0 A0A0Q9XCP6 Q7PQZ9 A0A182XI96 A0A182HT83 A0A182PA59 A0A182LFB8 A0A182NA84 Q171Y4 A0A336MIB4 A0A336MV82 A0A336MQG4 A0A182YKI4 U4TU65 N6UP99 U4U2T7 B4R207 A0A1Y1LJP8 A0A2J7RMT5 A0A2J7RMR6 A0A2J7RMR7 A0A2J7RMR9 A0A1Y1MHK6 A0A1Y1MG78 A0A139WFP4 A0A2H1WPZ4 A0A0C5KC89 A0A1Y1LQE6 A0A1S4F9W3 W8EA74
EC Number
3.1.1.-
Pubmed
19121390
26354079
22118469
26227816
25348373
25830018
+ More
25880816 24495485 26108605 17994087 15632085 23185243 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 18057021 12364791 14747013 17210077 20966253 17510324 25244985 23537049 28004739 18362917 19820115
25880816 24495485 26108605 17994087 15632085 23185243 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 18057021 12364791 14747013 17210077 20966253 17510324 25244985 23537049 28004739 18362917 19820115
EMBL
BABH01029942
BABH01029943
KQ459604
KPI92286.1
KQ460045
KPJ18206.1
+ More
AGBW02011379 OWR46748.1 NWSH01003327 PCG66554.1 GEYN01000029 JAV02100.1 ODYU01010411 SOQ55495.1 KY021870 AQY62759.1 JTDY01001671 KOB73199.1 GDHF01023319 GDHF01018887 GDHF01001753 JAI28995.1 JAI33427.1 JAI50561.1 GAKP01013206 JAC45746.1 GBXI01003993 JAD10299.1 KJ702184 AID61338.1 GAMC01013512 GAMC01013508 JAB93047.1 JRES01000232 KNC33141.1 GAMC01013509 JAB93046.1 CCAG010018501 CH916373 EDV94365.1 OUUW01000005 SPP81230.1 CH902617 EDV43991.2 CP012526 ALC46936.1 CM000070 EAL26945.4 EIM52339.2 KRS99673.1 CH479186 EDW38990.1 KRS99674.1 CM000160 KRK04137.1 BT100060 ACX61601.1 EDW98147.2 KRK04138.1 AJVK01007420 AJVK01007421 AJVK01007422 CH480815 EDW43339.1 CH954182 EDV54003.1 AXCN02000218 AY128457 AE014297 AAM75050.1 AAN13950.1 AXCM01000651 BT003545 AAO39549.1 KA648990 AFP63619.1 AAF56142.2 BT133184 AAN13949.1 AFA28425.1 AHN57486.1 CH964272 EDW83932.2 CH940650 EDW67362.1 KRF83261.1 KRF83259.1 GFDL01000479 JAV34566.1 GFDL01000520 JAV34525.1 GFDL01000503 JAV34542.1 GFDL01000461 JAV34584.1 CVRI01000070 CRL07130.1 CH933806 EDW16749.1 KRG02287.1 KRG02286.1 AAAB01008859 EAA08068.6 APCN01000421 APCN01000422 CH477443 EAT40802.2 UFQT01001349 SSX30154.1 UFQT01002094 SSX32653.1 SSX32652.1 KB631479 ERL84317.1 APGK01001751 KB734435 ENN83565.1 KB631477 KB631478 ERL84315.1 ERL84316.1 CM000364 EDX14064.1 GEZM01053959 GEZM01053958 JAV73894.1 NEVH01002552 PNF42132.1 PNF42130.1 PNF42131.1 PNF42133.1 GEZM01035171 JAV83376.1 GEZM01035172 JAV83375.1 KQ971352 KYB26709.1 ODYU01010136 SOQ55032.1 KP114631 AJP62543.1 GEZM01049757 JAV75869.1 KF849357 AHJ81320.1
AGBW02011379 OWR46748.1 NWSH01003327 PCG66554.1 GEYN01000029 JAV02100.1 ODYU01010411 SOQ55495.1 KY021870 AQY62759.1 JTDY01001671 KOB73199.1 GDHF01023319 GDHF01018887 GDHF01001753 JAI28995.1 JAI33427.1 JAI50561.1 GAKP01013206 JAC45746.1 GBXI01003993 JAD10299.1 KJ702184 AID61338.1 GAMC01013512 GAMC01013508 JAB93047.1 JRES01000232 KNC33141.1 GAMC01013509 JAB93046.1 CCAG010018501 CH916373 EDV94365.1 OUUW01000005 SPP81230.1 CH902617 EDV43991.2 CP012526 ALC46936.1 CM000070 EAL26945.4 EIM52339.2 KRS99673.1 CH479186 EDW38990.1 KRS99674.1 CM000160 KRK04137.1 BT100060 ACX61601.1 EDW98147.2 KRK04138.1 AJVK01007420 AJVK01007421 AJVK01007422 CH480815 EDW43339.1 CH954182 EDV54003.1 AXCN02000218 AY128457 AE014297 AAM75050.1 AAN13950.1 AXCM01000651 BT003545 AAO39549.1 KA648990 AFP63619.1 AAF56142.2 BT133184 AAN13949.1 AFA28425.1 AHN57486.1 CH964272 EDW83932.2 CH940650 EDW67362.1 KRF83261.1 KRF83259.1 GFDL01000479 JAV34566.1 GFDL01000520 JAV34525.1 GFDL01000503 JAV34542.1 GFDL01000461 JAV34584.1 CVRI01000070 CRL07130.1 CH933806 EDW16749.1 KRG02287.1 KRG02286.1 AAAB01008859 EAA08068.6 APCN01000421 APCN01000422 CH477443 EAT40802.2 UFQT01001349 SSX30154.1 UFQT01002094 SSX32653.1 SSX32652.1 KB631479 ERL84317.1 APGK01001751 KB734435 ENN83565.1 KB631477 KB631478 ERL84315.1 ERL84316.1 CM000364 EDX14064.1 GEZM01053959 GEZM01053958 JAV73894.1 NEVH01002552 PNF42132.1 PNF42130.1 PNF42131.1 PNF42133.1 GEZM01035171 JAV83376.1 GEZM01035172 JAV83375.1 KQ971352 KYB26709.1 ODYU01010136 SOQ55032.1 KP114631 AJP62543.1 GEZM01049757 JAV75869.1 KF849357 AHJ81320.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000007151
UP000218220
UP000037510
+ More
UP000069272 UP000037069 UP000095300 UP000092444 UP000078200 UP000091820 UP000001070 UP000268350 UP000092445 UP000007801 UP000092553 UP000001819 UP000092443 UP000075920 UP000075900 UP000008744 UP000002282 UP000092462 UP000192221 UP000001292 UP000008711 UP000075886 UP000000803 UP000075883 UP000095301 UP000007798 UP000008792 UP000075880 UP000183832 UP000009192 UP000075903 UP000007062 UP000076407 UP000075840 UP000075885 UP000075882 UP000075884 UP000008820 UP000076408 UP000030742 UP000019118 UP000000304 UP000235965 UP000007266
UP000069272 UP000037069 UP000095300 UP000092444 UP000078200 UP000091820 UP000001070 UP000268350 UP000092445 UP000007801 UP000092553 UP000001819 UP000092443 UP000075920 UP000075900 UP000008744 UP000002282 UP000092462 UP000192221 UP000001292 UP000008711 UP000075886 UP000000803 UP000075883 UP000095301 UP000007798 UP000008792 UP000075880 UP000183832 UP000009192 UP000075903 UP000007062 UP000076407 UP000075840 UP000075885 UP000075882 UP000075884 UP000008820 UP000076408 UP000030742 UP000019118 UP000000304 UP000235965 UP000007266
Interpro
IPR029058
AB_hydrolase
+ More
IPR002168 Lipase_GDXG_HIS_AS
IPR002018 CarbesteraseB
IPR019826 Carboxylesterase_B_AS
IPR019819 Carboxylesterase_B_CS
IPR035979 RBD_domain_sf
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR005829 Sugar_transporter_CS
IPR020846 MFS_dom
IPR036259 MFS_trans_sf
IPR005828 MFS_sugar_transport-like
IPR000997 Cholinesterase
IPR002168 Lipase_GDXG_HIS_AS
IPR002018 CarbesteraseB
IPR019826 Carboxylesterase_B_AS
IPR019819 Carboxylesterase_B_CS
IPR035979 RBD_domain_sf
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR005829 Sugar_transporter_CS
IPR020846 MFS_dom
IPR036259 MFS_trans_sf
IPR005828 MFS_sugar_transport-like
IPR000997 Cholinesterase
Gene 3D
CDD
ProteinModelPortal
H9J5Z3
A0A194PFX4
A0A194RQA4
A0A212EZ22
A0A2A4J4W9
A0A1L8D6Z3
+ More
A0A2H1WR02 A0A1U9X1X2 A0A0L7LCT1 A0A0K8URL8 A0A034VR46 A0A0A1XFZ8 A0A068F517 W8B3D5 A0A182F8C8 A0A0L0CLK8 W8BIG0 A0A1I8PWQ5 A0A1B0G3F2 A0A1A9UCU4 A0A1I8PWP1 A0A1I8PWQ3 A0A1I8PWQ1 A0A1A9X418 A0A1I8PWR0 A0A1I8PWN8 B4JRH5 A0A3B0K0B9 A0A1A9ZHC1 B3LY87 A0A0M5J219 Q29BQ2 I5AN04 A0A1A9Y8U9 A0A182VX55 A0A182R3N4 A0A0R3NF95 B4GPA7 A0A0R3NF77 A0A0R1E4Y1 D3DMF0 B4PN92 A0A0R1E9H0 A0A1B0F076 A0A1W4URC9 A0A1W4URD4 B4HFG1 B3P7B4 A0A1W4V4P9 A0A182QEU4 Q8MQQ0 A0A182MJA2 Q86P08 T1PLR5 Q9VCL6 Q8IMY6 A0A0B4LHM3 A0A1I8NHY5 B4NJK8 B4M0S0 A0A0Q9WFF2 A0A0Q9WRN5 A0A1Q3G476 A0A1Q3G3Z0 A0A1Q3G408 A0A182IRH2 A0A1Q3G467 A0A1J1J596 B4KA74 A0A182VPB0 A0A0Q9XCP6 Q7PQZ9 A0A182XI96 A0A182HT83 A0A182PA59 A0A182LFB8 A0A182NA84 Q171Y4 A0A336MIB4 A0A336MV82 A0A336MQG4 A0A182YKI4 U4TU65 N6UP99 U4U2T7 B4R207 A0A1Y1LJP8 A0A2J7RMT5 A0A2J7RMR6 A0A2J7RMR7 A0A2J7RMR9 A0A1Y1MHK6 A0A1Y1MG78 A0A139WFP4 A0A2H1WPZ4 A0A0C5KC89 A0A1Y1LQE6 A0A1S4F9W3 W8EA74
A0A2H1WR02 A0A1U9X1X2 A0A0L7LCT1 A0A0K8URL8 A0A034VR46 A0A0A1XFZ8 A0A068F517 W8B3D5 A0A182F8C8 A0A0L0CLK8 W8BIG0 A0A1I8PWQ5 A0A1B0G3F2 A0A1A9UCU4 A0A1I8PWP1 A0A1I8PWQ3 A0A1I8PWQ1 A0A1A9X418 A0A1I8PWR0 A0A1I8PWN8 B4JRH5 A0A3B0K0B9 A0A1A9ZHC1 B3LY87 A0A0M5J219 Q29BQ2 I5AN04 A0A1A9Y8U9 A0A182VX55 A0A182R3N4 A0A0R3NF95 B4GPA7 A0A0R3NF77 A0A0R1E4Y1 D3DMF0 B4PN92 A0A0R1E9H0 A0A1B0F076 A0A1W4URC9 A0A1W4URD4 B4HFG1 B3P7B4 A0A1W4V4P9 A0A182QEU4 Q8MQQ0 A0A182MJA2 Q86P08 T1PLR5 Q9VCL6 Q8IMY6 A0A0B4LHM3 A0A1I8NHY5 B4NJK8 B4M0S0 A0A0Q9WFF2 A0A0Q9WRN5 A0A1Q3G476 A0A1Q3G3Z0 A0A1Q3G408 A0A182IRH2 A0A1Q3G467 A0A1J1J596 B4KA74 A0A182VPB0 A0A0Q9XCP6 Q7PQZ9 A0A182XI96 A0A182HT83 A0A182PA59 A0A182LFB8 A0A182NA84 Q171Y4 A0A336MIB4 A0A336MV82 A0A336MQG4 A0A182YKI4 U4TU65 N6UP99 U4U2T7 B4R207 A0A1Y1LJP8 A0A2J7RMT5 A0A2J7RMR6 A0A2J7RMR7 A0A2J7RMR9 A0A1Y1MHK6 A0A1Y1MG78 A0A139WFP4 A0A2H1WPZ4 A0A0C5KC89 A0A1Y1LQE6 A0A1S4F9W3 W8EA74
PDB
5TYP
E-value=2.27443e-07,
Score=124
Ontologies
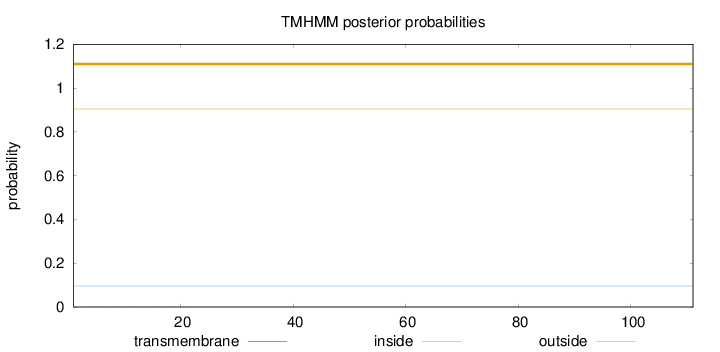
Topology
Length:
111
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00441
Exp number, first 60 AAs:
0.00441
Total prob of N-in:
0.09580
outside
1 - 111
Population Genetic Test Statistics
Pi
191.045369
Theta
134.393944
Tajima's D
1.265801
CLR
0
CSRT
0.729963501824909
Interpretation
Uncertain
