Pre Gene Modal
BGIBMGA004933
Annotation
NTF2-like_export_factor_1_[Bombyx_mori]
Full name
NTF2-related export protein
Alternative Name
p15
Location in the cell
Cytoplasmic Reliability : 1.703 Extracellular Reliability : 1.948
Sequence
CDS
ATGTCTGACGTAGCAGTAAAAAATGTTGATAGTGCTTGTGAGACAGCTGAAGAATTTACTAGAGTGTTTTACAAACAAGTAGACAACAGCCGCCATCTAACCTCAAAATTGTATTTGGATACTGGATTACTTGTTTGGAACGGCAATGGTATCAGTGGCAATGACAAAATACAGAAATTCTTGATGGATTTGCCTGCAAGTAACCATACGCTGAAAAGTTTAGATGCTCAACCAATATCTGATGTCTTGGTTGCAAATAAATTGACGTACCTCATCCAAGCTTGTGGAGATGTTACTTATCAAAACGAAGAAACAAAGAAACCATTTCAGCAAACTTTTCTCATAGTGGCAGTTGATGGGAAATGGAAGATAGCCTCGGATTGTTTCAGACTCCAAGTTCCTTATGGAAATTAA
Protein
MSDVAVKNVDSACETAEEFTRVFYKQVDNSRHLTSKLYLDTGLLVWNGNGISGNDKIQKFLMDLPASNHTLKSLDAQPISDVLVANKLTYLIQACGDVTYQNEETKKPFQQTFLIVAVDGKWKIASDCFRLQVPYGN
Summary
Description
Stimulator of protein export for NES-containing proteins (By similarity). Plays a role in the nuclear export of mRNA (PubMed:14729961). Also plays a role in the nuclear export of U1 snRNA, tRNA, and mRNA (By similarity).
Subunit
Forms a complex with RanGAP, Nup358/RanBP2 and sbr/Nxf1.
Keywords
Complete proteome
mRNA transport
Nucleus
Protein transport
Reference proteome
Transport
Feature
chain NTF2-related export protein
Uniprot
H9J5Z2
Q1HPZ2
A0A1E1WVF8
A0A0L7LP63
A0A2H1WQ09
S4PLN0
+ More
A0A194RLB3 A0A194PGW5 A0A2J7R807 D6WI27 A0A1W4WTG9 A0A034WMQ2 W8C9E6 A0A1Y1K4N1 A0A0K8WKN3 A0A067QPL3 B0XJC1 A0A1Q3G0R0 Q17KF0 A0A023EGI0 N6UQC4 A0A0L0CQB9 R4UL34 A0A1I8NLP4 U5EWI9 A0A1A9WIQ8 E2A3H6 A0A1I8MA70 A0A1B0G7P9 A0A1A9XP25 A0A1B0BNI3 A0A1A9V8C7 A0A084W4P5 A0A1A9ZWE1 A0A182UAJ3 A0A182XL16 Q5TQJ6 V9IIK2 A0A088ATI6 A0A2A3ET63 A0A182UQK6 A0A310SEF3 E9IZI1 A0A195CTU9 A0A158P476 A0A151WLK5 A0A151JR65 F4WIR6 A0A182MYW6 A0A232ERZ8 K7J767 A0A2R7WV06 A0A182L129 A0A182Q5S6 A0A2M4C2W5 A0A2M4C2Z9 A0A0N7Z8W2 A0A182JE74 A0A026W2A5 A0A1B0CAE5 A0A2M3ZK57 T1E8J4 W5JTA9 A0A1L8DGV6 A0A2M4AZI2 A0A069DN27 A0A2M4AZ73 T1IDZ0 A0A182W6T5 A0A182MLX5 B3ME81 A0A336MKH1 A0A182R557 A0A182PCL2 A0A0V0G602 A0A0M5J3X7 B4PA69 Q6XIJ1 A0A182Y3K1 B4L6L3 A0A336M6B3 A0A3B0J3K1 B3NPS6 B4NNM4 A0A0M4E8Y0 B4M7K3 B4JNI5 A0A0J7L4A8 Q293F8 B4GBK9 A0A1W4V013 A0A1B2AKG3 Q9V3H8 B4I8T5 B4QAZ7 A0A1B6EI12 A0A1B6GDK1 A0A1B6HSU5
A0A194RLB3 A0A194PGW5 A0A2J7R807 D6WI27 A0A1W4WTG9 A0A034WMQ2 W8C9E6 A0A1Y1K4N1 A0A0K8WKN3 A0A067QPL3 B0XJC1 A0A1Q3G0R0 Q17KF0 A0A023EGI0 N6UQC4 A0A0L0CQB9 R4UL34 A0A1I8NLP4 U5EWI9 A0A1A9WIQ8 E2A3H6 A0A1I8MA70 A0A1B0G7P9 A0A1A9XP25 A0A1B0BNI3 A0A1A9V8C7 A0A084W4P5 A0A1A9ZWE1 A0A182UAJ3 A0A182XL16 Q5TQJ6 V9IIK2 A0A088ATI6 A0A2A3ET63 A0A182UQK6 A0A310SEF3 E9IZI1 A0A195CTU9 A0A158P476 A0A151WLK5 A0A151JR65 F4WIR6 A0A182MYW6 A0A232ERZ8 K7J767 A0A2R7WV06 A0A182L129 A0A182Q5S6 A0A2M4C2W5 A0A2M4C2Z9 A0A0N7Z8W2 A0A182JE74 A0A026W2A5 A0A1B0CAE5 A0A2M3ZK57 T1E8J4 W5JTA9 A0A1L8DGV6 A0A2M4AZI2 A0A069DN27 A0A2M4AZ73 T1IDZ0 A0A182W6T5 A0A182MLX5 B3ME81 A0A336MKH1 A0A182R557 A0A182PCL2 A0A0V0G602 A0A0M5J3X7 B4PA69 Q6XIJ1 A0A182Y3K1 B4L6L3 A0A336M6B3 A0A3B0J3K1 B3NPS6 B4NNM4 A0A0M4E8Y0 B4M7K3 B4JNI5 A0A0J7L4A8 Q293F8 B4GBK9 A0A1W4V013 A0A1B2AKG3 Q9V3H8 B4I8T5 B4QAZ7 A0A1B6EI12 A0A1B6GDK1 A0A1B6HSU5
Pubmed
19121390
26227816
23622113
26354079
18362917
19820115
+ More
25348373 24495485 28004739 24845553 17510324 24945155 23537049 26108605 20798317 25315136 24438588 12364791 14747013 17210077 21282665 21347285 21719571 28648823 20075255 20966253 27129103 24508170 20920257 23761445 26334808 17994087 17550304 14525923 25244985 15632085 10567585 10731132 12537572 12537569 14729961 22936249
25348373 24495485 28004739 24845553 17510324 24945155 23537049 26108605 20798317 25315136 24438588 12364791 14747013 17210077 21282665 21347285 21719571 28648823 20075255 20966253 27129103 24508170 20920257 23761445 26334808 17994087 17550304 14525923 25244985 15632085 10567585 10731132 12537572 12537569 14729961 22936249
EMBL
BABH01029947
DQ443260
ABF51349.1
GDQN01006987
GDQN01000076
JAT84067.1
+ More
JAT90978.1 JTDY01000430 KOB77210.1 ODYU01010192 SOQ55149.1 GAIX01003945 JAA88615.1 KQ460045 KPJ18209.1 KQ459604 KPI92283.1 NEVH01006723 PNF36964.1 KQ971334 EEZ99700.2 GAKP01002126 JAC56826.1 GAMC01002224 JAC04332.1 GEZM01095244 JAV55441.1 GDHF01000695 JAI51619.1 KK853156 KDR10488.1 DS233478 EDS30158.1 GFDL01001662 JAV33383.1 CH477225 EAT47140.1 GAPW01005765 GAPW01005764 JAC07834.1 APGK01021186 KB740293 KB631672 ENN80942.1 ERL85269.1 JRES01000049 KNC34570.1 KC741025 AGM32849.1 GANO01001394 JAB58477.1 GL436428 EFN72040.1 CCAG010016890 JXJN01017413 ATLV01020364 KE525299 KFB45189.1 AAAB01008964 EAA12371.5 JR046098 AEY60134.1 KZ288185 PBC34935.1 KQ760672 OAD59444.1 GL767199 EFZ14021.1 KQ977279 KYN04116.1 ADTU01008694 KQ982959 KYQ48726.1 KQ978615 KYN29856.1 GL888177 EGI65862.1 NNAY01002531 OXU21125.1 AAZX01007534 KK855644 PTY23397.1 AXCN02000817 GGFJ01010493 MBW59634.1 GGFJ01010559 MBW59700.1 GDKW01002235 JAI54360.1 KK107485 EZA50128.1 AJWK01003706 AJWK01003707 GGFM01008148 MBW28899.1 GAMD01002718 JAA98872.1 ADMH02000146 GGFL01004616 ETN67612.1 MBW68794.1 GFDF01008479 JAV05605.1 GGFK01012801 MBW46122.1 GBGD01003426 JAC85463.1 GGFK01012773 MBW46094.1 ACPB03000233 AXCM01012135 CH902619 EDV35476.1 UFQT01001404 SSX30395.1 GECL01002725 JAP03399.1 CP012523 ALC39506.1 CM000158 EDW92395.1 AY231839 AAR09862.1 CH933812 EDW06009.1 UFQT01000450 SSX24493.1 OUUW01000001 SPP75947.1 CH954179 EDV56867.1 CH964282 EDW85963.1 CP012525 ALC43441.1 ALC44574.1 CH940653 EDW62770.1 CH916371 EDV92278.1 LBMM01000867 KMQ97329.1 CM000071 EAL24653.1 CH479181 EDW31304.1 KX531837 ANY27647.1 AF156959 AE013599 AY071250 CH480824 EDW57010.1 CM000362 CM002911 EDX08434.1 KMY96120.1 GECZ01032227 JAS37542.1 GECZ01009241 JAS60528.1 GECU01029952 JAS77754.1
JAT90978.1 JTDY01000430 KOB77210.1 ODYU01010192 SOQ55149.1 GAIX01003945 JAA88615.1 KQ460045 KPJ18209.1 KQ459604 KPI92283.1 NEVH01006723 PNF36964.1 KQ971334 EEZ99700.2 GAKP01002126 JAC56826.1 GAMC01002224 JAC04332.1 GEZM01095244 JAV55441.1 GDHF01000695 JAI51619.1 KK853156 KDR10488.1 DS233478 EDS30158.1 GFDL01001662 JAV33383.1 CH477225 EAT47140.1 GAPW01005765 GAPW01005764 JAC07834.1 APGK01021186 KB740293 KB631672 ENN80942.1 ERL85269.1 JRES01000049 KNC34570.1 KC741025 AGM32849.1 GANO01001394 JAB58477.1 GL436428 EFN72040.1 CCAG010016890 JXJN01017413 ATLV01020364 KE525299 KFB45189.1 AAAB01008964 EAA12371.5 JR046098 AEY60134.1 KZ288185 PBC34935.1 KQ760672 OAD59444.1 GL767199 EFZ14021.1 KQ977279 KYN04116.1 ADTU01008694 KQ982959 KYQ48726.1 KQ978615 KYN29856.1 GL888177 EGI65862.1 NNAY01002531 OXU21125.1 AAZX01007534 KK855644 PTY23397.1 AXCN02000817 GGFJ01010493 MBW59634.1 GGFJ01010559 MBW59700.1 GDKW01002235 JAI54360.1 KK107485 EZA50128.1 AJWK01003706 AJWK01003707 GGFM01008148 MBW28899.1 GAMD01002718 JAA98872.1 ADMH02000146 GGFL01004616 ETN67612.1 MBW68794.1 GFDF01008479 JAV05605.1 GGFK01012801 MBW46122.1 GBGD01003426 JAC85463.1 GGFK01012773 MBW46094.1 ACPB03000233 AXCM01012135 CH902619 EDV35476.1 UFQT01001404 SSX30395.1 GECL01002725 JAP03399.1 CP012523 ALC39506.1 CM000158 EDW92395.1 AY231839 AAR09862.1 CH933812 EDW06009.1 UFQT01000450 SSX24493.1 OUUW01000001 SPP75947.1 CH954179 EDV56867.1 CH964282 EDW85963.1 CP012525 ALC43441.1 ALC44574.1 CH940653 EDW62770.1 CH916371 EDV92278.1 LBMM01000867 KMQ97329.1 CM000071 EAL24653.1 CH479181 EDW31304.1 KX531837 ANY27647.1 AF156959 AE013599 AY071250 CH480824 EDW57010.1 CM000362 CM002911 EDX08434.1 KMY96120.1 GECZ01032227 JAS37542.1 GECZ01009241 JAS60528.1 GECU01029952 JAS77754.1
Proteomes
UP000005204
UP000037510
UP000053240
UP000053268
UP000235965
UP000007266
+ More
UP000192223 UP000027135 UP000002320 UP000008820 UP000019118 UP000030742 UP000037069 UP000095300 UP000091820 UP000000311 UP000095301 UP000092444 UP000092443 UP000092460 UP000078200 UP000030765 UP000092445 UP000075902 UP000076407 UP000007062 UP000005203 UP000242457 UP000075903 UP000078542 UP000005205 UP000075809 UP000078492 UP000007755 UP000075884 UP000215335 UP000002358 UP000075882 UP000075886 UP000075880 UP000053097 UP000092461 UP000000673 UP000015103 UP000075920 UP000075883 UP000007801 UP000075900 UP000075885 UP000092553 UP000002282 UP000076408 UP000009192 UP000268350 UP000008711 UP000007798 UP000008792 UP000001070 UP000036403 UP000001819 UP000008744 UP000192221 UP000000803 UP000001292 UP000000304
UP000192223 UP000027135 UP000002320 UP000008820 UP000019118 UP000030742 UP000037069 UP000095300 UP000091820 UP000000311 UP000095301 UP000092444 UP000092443 UP000092460 UP000078200 UP000030765 UP000092445 UP000075902 UP000076407 UP000007062 UP000005203 UP000242457 UP000075903 UP000078542 UP000005205 UP000075809 UP000078492 UP000007755 UP000075884 UP000215335 UP000002358 UP000075882 UP000075886 UP000075880 UP000053097 UP000092461 UP000000673 UP000015103 UP000075920 UP000075883 UP000007801 UP000075900 UP000075885 UP000092553 UP000002282 UP000076408 UP000009192 UP000268350 UP000008711 UP000007798 UP000008792 UP000001070 UP000036403 UP000001819 UP000008744 UP000192221 UP000000803 UP000001292 UP000000304
Pfam
PF02136 NTF2
SUPFAM
SSF54427
SSF54427
CDD
ProteinModelPortal
H9J5Z2
Q1HPZ2
A0A1E1WVF8
A0A0L7LP63
A0A2H1WQ09
S4PLN0
+ More
A0A194RLB3 A0A194PGW5 A0A2J7R807 D6WI27 A0A1W4WTG9 A0A034WMQ2 W8C9E6 A0A1Y1K4N1 A0A0K8WKN3 A0A067QPL3 B0XJC1 A0A1Q3G0R0 Q17KF0 A0A023EGI0 N6UQC4 A0A0L0CQB9 R4UL34 A0A1I8NLP4 U5EWI9 A0A1A9WIQ8 E2A3H6 A0A1I8MA70 A0A1B0G7P9 A0A1A9XP25 A0A1B0BNI3 A0A1A9V8C7 A0A084W4P5 A0A1A9ZWE1 A0A182UAJ3 A0A182XL16 Q5TQJ6 V9IIK2 A0A088ATI6 A0A2A3ET63 A0A182UQK6 A0A310SEF3 E9IZI1 A0A195CTU9 A0A158P476 A0A151WLK5 A0A151JR65 F4WIR6 A0A182MYW6 A0A232ERZ8 K7J767 A0A2R7WV06 A0A182L129 A0A182Q5S6 A0A2M4C2W5 A0A2M4C2Z9 A0A0N7Z8W2 A0A182JE74 A0A026W2A5 A0A1B0CAE5 A0A2M3ZK57 T1E8J4 W5JTA9 A0A1L8DGV6 A0A2M4AZI2 A0A069DN27 A0A2M4AZ73 T1IDZ0 A0A182W6T5 A0A182MLX5 B3ME81 A0A336MKH1 A0A182R557 A0A182PCL2 A0A0V0G602 A0A0M5J3X7 B4PA69 Q6XIJ1 A0A182Y3K1 B4L6L3 A0A336M6B3 A0A3B0J3K1 B3NPS6 B4NNM4 A0A0M4E8Y0 B4M7K3 B4JNI5 A0A0J7L4A8 Q293F8 B4GBK9 A0A1W4V013 A0A1B2AKG3 Q9V3H8 B4I8T5 B4QAZ7 A0A1B6EI12 A0A1B6GDK1 A0A1B6HSU5
A0A194RLB3 A0A194PGW5 A0A2J7R807 D6WI27 A0A1W4WTG9 A0A034WMQ2 W8C9E6 A0A1Y1K4N1 A0A0K8WKN3 A0A067QPL3 B0XJC1 A0A1Q3G0R0 Q17KF0 A0A023EGI0 N6UQC4 A0A0L0CQB9 R4UL34 A0A1I8NLP4 U5EWI9 A0A1A9WIQ8 E2A3H6 A0A1I8MA70 A0A1B0G7P9 A0A1A9XP25 A0A1B0BNI3 A0A1A9V8C7 A0A084W4P5 A0A1A9ZWE1 A0A182UAJ3 A0A182XL16 Q5TQJ6 V9IIK2 A0A088ATI6 A0A2A3ET63 A0A182UQK6 A0A310SEF3 E9IZI1 A0A195CTU9 A0A158P476 A0A151WLK5 A0A151JR65 F4WIR6 A0A182MYW6 A0A232ERZ8 K7J767 A0A2R7WV06 A0A182L129 A0A182Q5S6 A0A2M4C2W5 A0A2M4C2Z9 A0A0N7Z8W2 A0A182JE74 A0A026W2A5 A0A1B0CAE5 A0A2M3ZK57 T1E8J4 W5JTA9 A0A1L8DGV6 A0A2M4AZI2 A0A069DN27 A0A2M4AZ73 T1IDZ0 A0A182W6T5 A0A182MLX5 B3ME81 A0A336MKH1 A0A182R557 A0A182PCL2 A0A0V0G602 A0A0M5J3X7 B4PA69 Q6XIJ1 A0A182Y3K1 B4L6L3 A0A336M6B3 A0A3B0J3K1 B3NPS6 B4NNM4 A0A0M4E8Y0 B4M7K3 B4JNI5 A0A0J7L4A8 Q293F8 B4GBK9 A0A1W4V013 A0A1B2AKG3 Q9V3H8 B4I8T5 B4QAZ7 A0A1B6EI12 A0A1B6GDK1 A0A1B6HSU5
PDB
6E5U
E-value=3.02329e-24,
Score=270
Ontologies
PATHWAY
GO
Topology
Subcellular location
Nucleus
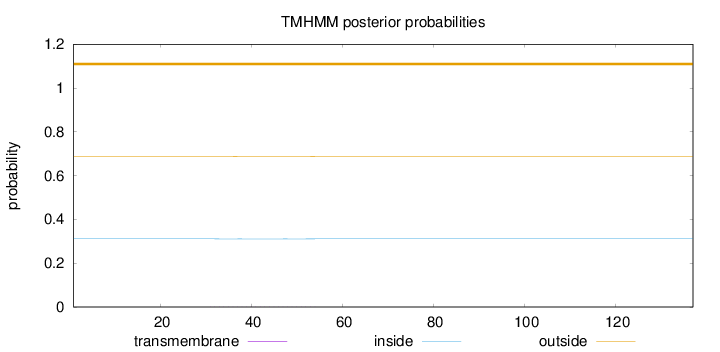
Length:
137
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02331
Exp number, first 60 AAs:
0.02171
Total prob of N-in:
0.31123
outside
1 - 137
Population Genetic Test Statistics
Pi
327.949289
Theta
223.141051
Tajima's D
1.479047
CLR
84.607205
CSRT
0.784110794460277
Interpretation
Uncertain
