Gene
KWMTBOMO15159 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA004932
Annotation
PREDICTED:_ribosome-binding_protein_1_isoform_X1_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.554 Nuclear Reliability : 1.141
Sequence
CDS
ATGGAGCTGCAAGCATTACTGGTTCTGGGAGGATTGGGGGTGGCTGGCATAGCAGTTCTCCTATTGATGGGACTGTTCTCAGCCTCAGGCACTAGCTATGAAGAGGCTATTGCACAACAACGTAAAGCAGCTACTGAGTTGTTGGCATTAGCAGAAAACAAAAGCAAGTCGAAGAAAAATGTGAAGAAGAAGGATAAGAAGCTGGCCAAGAAAGAGAAGAGCAAGGAGAACTCTGGTACAACAGGCAGTGAAATCGACAGTGAGGCCCCAGCTGAGAGTGGTCTAGAAGATGACCCCACTCCACCAACTAAGGGACATGTCGAATTCTCCCCACCTGTTGTTGTTGATGTACCCAGGGATGCACCCCCTAATGTTAAGATACGCAAACGTGGCAAGGATCCTAAGGTAAAGCCGATTCTGTTGAACAAAGAGGATCCGAGTTGCGTTAGTGATCCGAGCACGGTCCCAGTGGCCGCCAATACCACTGTCGCTAACCATTTTGAGGAAATGCATCCAAAAGATGAAATAGAATTGATGCAATCGGCCCTATCAACTGAAAAAGTCGTTGAAAAAGTTGAGGAACCTGTTGACAAGAAGGAAGCAAAGCCTACAAAACCTCAAAAAACTGGCAAAGGTGCTCCTAAAGCAACTTCATCCCCTGAGCTTAAAGAAGAAACAAAAGAAAAAAACAACACTGGGGACGCACCCAAGGAGCAACGTAAAGGTAAAAAAGAGAAGAAGCCTCATGATGAAGCAGCGGAAGTAGCACAACAGGAGATTGTTCCACCATTGAACATACCACAGCCGACTGAGCTAACCACCGAAAAACTTCTGAAACAAGCATTAGTTGTCGCACCTCCTGCGCCGTCGCCTCCAGCTGGCAAGGGGAAGAAGAAAAAACAAGAACCTAATGTCCTCACGCTCATGGTTGGCGACCACGGTGGCGTAAACGTTGGCGAGCTGGTGCGCGTGGTCCGCGAGGCTTCGCTCTCTCGTACAGAGATACAGATCCTGACCGACGCGTTGCTCGACAAACACCATGATCCTCTACCCGAACATTCCGAATGGTCCGAGGGCCCAAATGATCCAGTTCAGAAATTGAAGAAGCAGTTAGCCGACAAAGAAAAGGCGCTGGCCGATGAAGTTGAAGCTTCCCAAGCTTTGCAAGCCAAATTGAAGGAATTACGAACGAGCCTGAACGCAGAGCGCAGCCGGGCGAGCGGCGCCGTGCGCACGCTGGAGGCGGAGCGCGCCCGCCACGAGCAGGCCGCGGCCGCCGCGCGCGTCGACCTGCACACGCACCAGGCGCGCCTGCAGCGGCTCCTCGACGACAACCACCAGCTCGCACAGGAGAAGCTGCACTTGCAATCGCAATTGAATGCAGAAGCGGAAGTTCAAGCTCAACGAGTCCAGATGGAGGTGCACATTAAGAGATTAACAGAGTCGGAGGCGGCGCTGGTGCAGCAGCTGGCGGGGCTGCAGAGCGAGGCGGCCGCGCACGCGCACGACGCCCGCCAGGCGCGCCTCGAGCTGGGCGCCGCCCGCGACGCGCTGCTCATGGCGCAGCAGCACGCCGCCGAACTGGCGCAACAACTGCAGGATGCAAACCGTGCATACACCGAACTAGAGCAGCACTCTCAACAAGAAAGAAGGCAATTACAGGAACGACTCGCTGAGCTTCAAAATGAAGTCCAGAGATTGGCGAATCATGCTCAAACAGTCGAAGCGAACACAGAGAAACTAAAACAAGAAAGTGAAAAACAAAAACAACAGCTATCCAATGAGATCAAAATAATAGGGGAGCAGCTGTCCGCGCGCGAGGCCGAGCTCGCTGAGCTCAAACAGATAAAGGCGGTGCCCGCACAGAACGGTTTGACGGCGGATGACAATGAACAAAAGATAAGCAAATCGGAGTTCGCGAAGGTGGAGAGCGTGGTGGCGGCGCTGCGCAGCGAGCTGGAGTCTGCGCAGGGCAGCAGCCGCGAGCAGCGCGACCAGCTCGAGCGGCTGCGCGACGAGCTGCACCGCCACAAGCACAAGAACGACGAGTTGCGAACTAAAAACTGGAAAGTAATGGAGGCGTTACAGTCTGCCGAAAAGGCTCTCCAGTCGAGAACGGCAAGCGCGTTGCCGGCCCAAGACTCGTTGCGTGAGGCAGTGGCGAAGGCGCAGGAAACGCAACTAGCTGAAGTGGCCGCTGTGCTACGCGCCGCCTGCCCCGCCGCCGCGCCCCATGACCCCTGCGCAGACCTCGCCTGGCTGAGAACCTTCGCCGACAACCTCGGACAATTGCTGCTGCAGCAGAAGGACTTGCTGCTCAAGGAGACTGAGAAGCAGGCGACAGTGGTCACGGCGCCGCCCTCTGACGACCGACTCAAGGAGCTGGCCGATCAGAACGAACACCTCCAAAGTCTCGTCGACAAATACAAGACGATCATAGACGACACGGAGGGCGTGTTAAGCCGGCTACAGCAAAACGTGACGACAGAGGAACAGCGATGGGCGCAACAACTCGCCGACAAACAGCGGGAACTCGATGAACTGCGTCAGAGAACTGTTTCGCAGATGCAGAATAAGATTGATTCGTTGCAAGAAGAACTGGCGCAGGCCAAATCAGCGCGACACAATCACACGTTCGCCGACGCCGAGCGTCTCGCCGAGGAAAGACTGATGGCGGGTTTGTCTGATAAACATACGGATATCAGCAACGGACCGTTACAGCTCGGCTTGGAAAATAAATAG
Protein
MELQALLVLGGLGVAGIAVLLLMGLFSASGTSYEEAIAQQRKAATELLALAENKSKSKKNVKKKDKKLAKKEKSKENSGTTGSEIDSEAPAESGLEDDPTPPTKGHVEFSPPVVVDVPRDAPPNVKIRKRGKDPKVKPILLNKEDPSCVSDPSTVPVAANTTVANHFEEMHPKDEIELMQSALSTEKVVEKVEEPVDKKEAKPTKPQKTGKGAPKATSSPELKEETKEKNNTGDAPKEQRKGKKEKKPHDEAAEVAQQEIVPPLNIPQPTELTTEKLLKQALVVAPPAPSPPAGKGKKKKQEPNVLTLMVGDHGGVNVGELVRVVREASLSRTEIQILTDALLDKHHDPLPEHSEWSEGPNDPVQKLKKQLADKEKALADEVEASQALQAKLKELRTSLNAERSRASGAVRTLEAERARHEQAAAAARVDLHTHQARLQRLLDDNHQLAQEKLHLQSQLNAEAEVQAQRVQMEVHIKRLTESEAALVQQLAGLQSEAAAHAHDARQARLELGAARDALLMAQQHAAELAQQLQDANRAYTELEQHSQQERRQLQERLAELQNEVQRLANHAQTVEANTEKLKQESEKQKQQLSNEIKIIGEQLSAREAELAELKQIKAVPAQNGLTADDNEQKISKSEFAKVESVVAALRSELESAQGSSREQRDQLERLRDELHRHKHKNDELRTKNWKVMEALQSAEKALQSRTASALPAQDSLREAVAKAQETQLAEVAAVLRAACPAAAPHDPCADLAWLRTFADNLGQLLLQQKDLLLKETEKQATVVTAPPSDDRLKELADQNEHLQSLVDKYKTIIDDTEGVLSRLQQNVTTEEQRWAQQLADKQRELDELRQRTVSQMQNKIDSLQEELAQAKSARHNHTFADAERLAEERLMAGLSDKHTDISNGPLQLGLENK
Summary
Uniprot
Pubmed
EMBL
Proteomes
PRIDE
Interpro
IPR040248
RRBP1
ProteinModelPortal
PDB
4RH7
E-value=0.0358889,
Score=90
Ontologies
GO
PANTHER
Topology
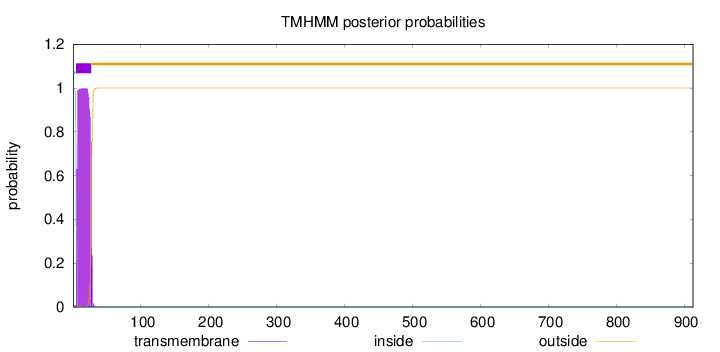
Length:
913
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.23018
Exp number, first 60 AAs:
22.22997
Total prob of N-in:
0.99804
POSSIBLE N-term signal
sequence
inside
1 - 4
TMhelix
5 - 27
outside
28 - 913
Population Genetic Test Statistics
Pi
238.058332
Theta
197.780312
Tajima's D
0.684386
CLR
0.200133
CSRT
0.571271436428179
Interpretation
Uncertain
