Gene
KWMTBOMO15157
Pre Gene Modal
BGIBMGA004931
Annotation
PREDICTED:_dimethyladenosine_transferase_2?_mitochondrial_[Bombyx_mori]
Full name
rRNA adenine N(6)-methyltransferase
+ More
Dimethyladenosine transferase 2, mitochondrial
Pyruvate kinase
Dimethyladenosine transferase 2, mitochondrial
Pyruvate kinase
Alternative Name
Mitochondrial 12S rRNA dimethylase 2
Mitochondrial transcription factor B2
S-adenosylmethionine-6-N', N'-adenosyl(rRNA) dimethyltransferase 2
d-mtTFB2
Mitochondrial transcription factor B2
S-adenosylmethionine-6-N', N'-adenosyl(rRNA) dimethyltransferase 2
d-mtTFB2
Location in the cell
Cytoplasmic Reliability : 1.018 Mitochondrial Reliability : 1.267 Nuclear Reliability : 1.33
Sequence
CDS
ATGCGTCACAAATTAAATAAACTGAGTGAAAAACAGCCGAAATTGGCCGAAGATGTTCTCAAGTTTTTAAATAGTTCACCAGAATACAATGGTTTGAAAGAACAACTTCCTACGTCATTACTTAGAAAATATAAAGCACCTGAAAGTTCACCATTGATTGAGGTTAACCCTGGCTTGGGTATTCTTACTGAAAATATTCTAAAATGTCAAAATAACAAGATATACTTATTTGAACAGTCTAATCATTTTTCTCCATACTTGGATGACTTACAACAAAAATACCCAGGCAGGATCTCATTCAAAATAGCAGATTTTTTTGGAATGTGGAAATTAGCATTCAAAGATAAAATGGATGATGGAAACAGAATTCAGGAATTGCTAGGAGATTTATCTACTACTGACAAAAATAGGACTGTCAAGATACTGGGATCAATGCCAGGACTATCTTTCATGAAACATTTGATTTACAATGTATTATTCCATAACACTACAAATCAGCTCGGTAGACCTGATCTGTTTATAACAATGCCTGTACATCATTATGAGTTTCTTACAGATAAGGAGGTTCAGCAATCAAAACAGAAATCTGTGCCAACCTTATTCCAGCTTTTGTTTGATTGCAAAACATTAACAACGGTTACAAAAGCCCATTTTCTGCCATGGACATTGAGTAATAGCAAACACCATTCTGTGCTGAATGAACACAACCTGTATTTGGTAAATATAACACAAAAGGAAAAACTACCATGTCCAGCAGAATATTTGCCATTGTTATGGTATTTTTTCAAACCACAAATGTTCTCGAAATCGACGAGAGTTATTCCGATGTTGGAGCAATGGATACCAGGGTGCGGAGTGTGGCTTATAACTGGACAGGATCCACCGGACAGCAATAAATTATTATCCCCAGGAAAAGATGACGCGTCACTGCCACATATGACCATATTTACAGAGTTTGGTGATCTTAATTTACAACAAAAAATTACTGTTTTCAAAAGGTACATTTTCTATATATCATAA
Protein
MRHKLNKLSEKQPKLAEDVLKFLNSSPEYNGLKEQLPTSLLRKYKAPESSPLIEVNPGLGILTENILKCQNNKIYLFEQSNHFSPYLDDLQQKYPGRISFKIADFFGMWKLAFKDKMDDGNRIQELLGDLSTTDKNRTVKILGSMPGLSFMKHLIYNVLFHNTTNQLGRPDLFITMPVHHYEFLTDKEVQQSKQKSVPTLFQLLFDCKTLTTVTKAHFLPWTLSNSKHHSVLNEHNLYLVNITQKEKLPCPAEYLPLLWYFFKPQMFSKSTRVIPMLEQWIPGCGVWLITGQDPPDSNKLLSPGKDDASLPHMTIFTEFGDLNLQQKITVFKRYIFYIS
Summary
Description
Probable S-adenosyl-L-methionine-dependent methyltransferase which specifically dimethylates mitochondrial 12S rRNA at the conserved stem loop. Also required for basal transcription of mitochondrial DNA. Also regulates mitochondrial DNA copy number. Stimulates transcription independently of the methyltransferase activity.
Catalytic Activity
ATP + pyruvate = ADP + H(+) + phosphoenolpyruvate
Similarity
Belongs to the class I-like SAM-binding methyltransferase superfamily. rRNA adenine N(6)-methyltransferase family.
Belongs to the class I-like SAM-binding methyltransferase superfamily. rRNA adenine N(6)-methyltransferase family. KsgA subfamily.
Belongs to the pyruvate kinase family.
Belongs to the class I-like SAM-binding methyltransferase superfamily. rRNA adenine N(6)-methyltransferase family. KsgA subfamily.
Belongs to the pyruvate kinase family.
Keywords
Complete proteome
Methyltransferase
Mitochondrion
Reference proteome
RNA-binding
rRNA processing
S-adenosyl-L-methionine
Transcription
Transcription regulation
Transferase
Transit peptide
Feature
chain Dimethyladenosine transferase 2, mitochondrial
Uniprot
A0A2H1WV65
A0A1E1WHZ5
A0A1E1W153
A0A194PMD7
A0A194RKZ2
A0A212FE22
+ More
A0A034V410 A0A0A1X1H7 A0A034V534 A0A0K8VXA9 B4JSL0 A0A1I8QD22 W8AS10 B4KCG3 A0A1A9XNN1 A0A1B0DNZ7 B4NFL8 A0A1L8E141 B4M688 A0A2J7QNZ3 A0A0K8VKI4 B3P4M6 A0A1B0BK93 A0A1A9ZQ04 B4GZ09 A0A1A9VZA8 A0A067QQU3 A0A1I8MMU9 B4QVV5 A0A3B0K392 A0A1J1HU80 A0A1B0FHF6 Q9VH38 B4HJA9 Q29B70 B3M1F0 A0A2J7QNZ6 B4PVI5 A0A1B3PDF8 A0A0M5J4W3 A0A0L0BKZ3 A0A1W4W0K1 A0A1A9VNU8 A0A1B3PDB3 A0A023EYN0 A0A1Q3F632 A0A182H1Q2 A0A0K8U2I3 A0A336MXX8 J9JY26 A0A1B6D0A2 A0A1B6E0S9 A0A2S2QM08 A0A1S4FWI2 Q16LP3 A0A224XS25 A0A182QP86 A0A182TAI1 A0A182Y3T3 A0A026W748 E2BCN1 A0A0J7NQY4 Q16F37 A0A2H8TWS3 A0A182NRT5 A0A139WEP0 A0A2M4AWE9 A0A2M4AW36 A0A182JG70 W5JWF5 A0A182MR20 A0A182FP48 E2AVH6 N6SSI5 A0A182VZM2 A0A1Y1KCK8 A0A195C9S0 A0A1B6IKN4 A0A182TXU5 A0A182LA32 Q7PY27 A0A182PSI6 A0A182UTL6 A0A182HLN8 A0A182JU81 A0A1B6L546 A0A151IXH4 A0A0L7R1J5 A0A2M4BSH0 A0A2R7W7F0 A0A2M4BTB2 A0A2M4CNV7 A0A293N6V4 A0A084VLC0 A0A088A0E7 A0A182WST4 A0A0M8ZUN4 A0A0A9W2A9
A0A034V410 A0A0A1X1H7 A0A034V534 A0A0K8VXA9 B4JSL0 A0A1I8QD22 W8AS10 B4KCG3 A0A1A9XNN1 A0A1B0DNZ7 B4NFL8 A0A1L8E141 B4M688 A0A2J7QNZ3 A0A0K8VKI4 B3P4M6 A0A1B0BK93 A0A1A9ZQ04 B4GZ09 A0A1A9VZA8 A0A067QQU3 A0A1I8MMU9 B4QVV5 A0A3B0K392 A0A1J1HU80 A0A1B0FHF6 Q9VH38 B4HJA9 Q29B70 B3M1F0 A0A2J7QNZ6 B4PVI5 A0A1B3PDF8 A0A0M5J4W3 A0A0L0BKZ3 A0A1W4W0K1 A0A1A9VNU8 A0A1B3PDB3 A0A023EYN0 A0A1Q3F632 A0A182H1Q2 A0A0K8U2I3 A0A336MXX8 J9JY26 A0A1B6D0A2 A0A1B6E0S9 A0A2S2QM08 A0A1S4FWI2 Q16LP3 A0A224XS25 A0A182QP86 A0A182TAI1 A0A182Y3T3 A0A026W748 E2BCN1 A0A0J7NQY4 Q16F37 A0A2H8TWS3 A0A182NRT5 A0A139WEP0 A0A2M4AWE9 A0A2M4AW36 A0A182JG70 W5JWF5 A0A182MR20 A0A182FP48 E2AVH6 N6SSI5 A0A182VZM2 A0A1Y1KCK8 A0A195C9S0 A0A1B6IKN4 A0A182TXU5 A0A182LA32 Q7PY27 A0A182PSI6 A0A182UTL6 A0A182HLN8 A0A182JU81 A0A1B6L546 A0A151IXH4 A0A0L7R1J5 A0A2M4BSH0 A0A2R7W7F0 A0A2M4BTB2 A0A2M4CNV7 A0A293N6V4 A0A084VLC0 A0A088A0E7 A0A182WST4 A0A0M8ZUN4 A0A0A9W2A9
EC Number
2.1.1.-
2.7.1.40
2.7.1.40
Pubmed
26354079
22118469
25348373
25830018
17994087
24495485
+ More
24845553 25315136 15060065 10731132 12537572 12537569 15632085 17550304 26108605 25474469 26483478 17510324 25244985 24508170 30249741 20798317 18362917 19820115 20920257 23761445 23537049 28004739 20966253 12364791 14747013 17210077 24438588 25401762 26823975
24845553 25315136 15060065 10731132 12537572 12537569 15632085 17550304 26108605 25474469 26483478 17510324 25244985 24508170 30249741 20798317 18362917 19820115 20920257 23761445 23537049 28004739 20966253 12364791 14747013 17210077 24438588 25401762 26823975
EMBL
ODYU01011303
SOQ56953.1
GDQN01004517
JAT86537.1
GDQN01010392
JAT80662.1
+ More
KQ459604 KPI92280.1 KQ460045 KPJ18212.1 AGBW02008977 OWR52015.1 GAKP01022065 JAC36887.1 GBXI01009789 JAD04503.1 GAKP01022067 JAC36885.1 GDHF01008798 JAI43516.1 CH916373 EDV94750.1 GAMC01017704 GAMC01017701 GAMC01017698 GAMC01017694 JAB88857.1 CH933806 EDW14782.1 AJVK01017930 AJVK01017931 CH964251 EDW83085.1 GFDF01001700 JAV12384.1 CH940652 EDW59164.1 NEVH01012098 PNF30306.1 GDHF01012923 JAI39391.1 CH954181 EDV49679.1 JXJN01015848 CH479198 EDW28027.1 KK853074 KDR11763.1 CM000364 EDX13535.1 OUUW01000005 SPP80429.1 CVRI01000015 CRK90054.1 CCAG010006669 AY508412 AE014297 AY060616 CH480815 EDW42781.1 CM000070 EAL27128.2 CH902617 EDV42177.1 PNF30309.1 CM000160 EDW96758.1 KU659969 AOG17767.1 CP012526 ALC46461.1 JRES01001712 KNC20603.1 KU659916 AOG17715.1 GBBI01004470 JAC14242.1 GFDL01012117 JAV22928.1 JXUM01023189 JXUM01023190 KQ560654 KXJ81422.1 GDHF01031603 JAI20711.1 UFQS01003764 UFQT01003764 SSX15920.1 SSX35262.1 ABLF02022843 GEDC01018166 JAS19132.1 GEDC01005774 JAS31524.1 GGMS01008989 MBY78192.1 CH477898 EAT35245.1 GFTR01005483 JAW10943.1 AXCN02001148 KK107364 QOIP01000007 EZA51932.1 RLU20370.1 GL447336 EFN86541.1 LBMM01002400 KMQ94875.1 CH478516 EAT32847.1 GFXV01006902 MBW18707.1 KQ971354 KYB26311.1 GGFK01011597 MBW44918.1 GGFK01011654 MBW44975.1 ADMH02000156 ETN67550.1 AXCM01000026 GL443112 EFN62571.1 APGK01058767 KB741291 KB631759 ENN70609.1 ERL85851.1 GEZM01088526 JAV58131.1 KQ978068 KYM97609.1 GECU01020227 JAS87479.1 AAAB01008987 EAA00845.4 APCN01000909 GEBQ01021124 JAT18853.1 KQ980832 KYN12397.1 KQ414667 KOC64740.1 GGFJ01006868 MBW56009.1 KK854345 PTY14910.1 GGFJ01006867 MBW56008.1 GGFL01002771 MBW66949.1 GFWV01023211 MAA47938.1 ATLV01014443 KE524972 KFB38764.1 KQ435863 KOX70356.1 GBHO01042053 GBRD01012169 GDHC01010996 JAG01551.1 JAG53655.1 JAQ07633.1
KQ459604 KPI92280.1 KQ460045 KPJ18212.1 AGBW02008977 OWR52015.1 GAKP01022065 JAC36887.1 GBXI01009789 JAD04503.1 GAKP01022067 JAC36885.1 GDHF01008798 JAI43516.1 CH916373 EDV94750.1 GAMC01017704 GAMC01017701 GAMC01017698 GAMC01017694 JAB88857.1 CH933806 EDW14782.1 AJVK01017930 AJVK01017931 CH964251 EDW83085.1 GFDF01001700 JAV12384.1 CH940652 EDW59164.1 NEVH01012098 PNF30306.1 GDHF01012923 JAI39391.1 CH954181 EDV49679.1 JXJN01015848 CH479198 EDW28027.1 KK853074 KDR11763.1 CM000364 EDX13535.1 OUUW01000005 SPP80429.1 CVRI01000015 CRK90054.1 CCAG010006669 AY508412 AE014297 AY060616 CH480815 EDW42781.1 CM000070 EAL27128.2 CH902617 EDV42177.1 PNF30309.1 CM000160 EDW96758.1 KU659969 AOG17767.1 CP012526 ALC46461.1 JRES01001712 KNC20603.1 KU659916 AOG17715.1 GBBI01004470 JAC14242.1 GFDL01012117 JAV22928.1 JXUM01023189 JXUM01023190 KQ560654 KXJ81422.1 GDHF01031603 JAI20711.1 UFQS01003764 UFQT01003764 SSX15920.1 SSX35262.1 ABLF02022843 GEDC01018166 JAS19132.1 GEDC01005774 JAS31524.1 GGMS01008989 MBY78192.1 CH477898 EAT35245.1 GFTR01005483 JAW10943.1 AXCN02001148 KK107364 QOIP01000007 EZA51932.1 RLU20370.1 GL447336 EFN86541.1 LBMM01002400 KMQ94875.1 CH478516 EAT32847.1 GFXV01006902 MBW18707.1 KQ971354 KYB26311.1 GGFK01011597 MBW44918.1 GGFK01011654 MBW44975.1 ADMH02000156 ETN67550.1 AXCM01000026 GL443112 EFN62571.1 APGK01058767 KB741291 KB631759 ENN70609.1 ERL85851.1 GEZM01088526 JAV58131.1 KQ978068 KYM97609.1 GECU01020227 JAS87479.1 AAAB01008987 EAA00845.4 APCN01000909 GEBQ01021124 JAT18853.1 KQ980832 KYN12397.1 KQ414667 KOC64740.1 GGFJ01006868 MBW56009.1 KK854345 PTY14910.1 GGFJ01006867 MBW56008.1 GGFL01002771 MBW66949.1 GFWV01023211 MAA47938.1 ATLV01014443 KE524972 KFB38764.1 KQ435863 KOX70356.1 GBHO01042053 GBRD01012169 GDHC01010996 JAG01551.1 JAG53655.1 JAQ07633.1
Proteomes
UP000053268
UP000053240
UP000007151
UP000001070
UP000095300
UP000009192
+ More
UP000092443 UP000092462 UP000007798 UP000008792 UP000235965 UP000008711 UP000092460 UP000092445 UP000008744 UP000091820 UP000027135 UP000095301 UP000000304 UP000268350 UP000183832 UP000092444 UP000000803 UP000001292 UP000001819 UP000007801 UP000002282 UP000092553 UP000037069 UP000192221 UP000078200 UP000069940 UP000249989 UP000007819 UP000008820 UP000075886 UP000075901 UP000076408 UP000053097 UP000279307 UP000008237 UP000036403 UP000075884 UP000007266 UP000075880 UP000000673 UP000075883 UP000069272 UP000000311 UP000019118 UP000030742 UP000075920 UP000078542 UP000075902 UP000075882 UP000007062 UP000075885 UP000075903 UP000075840 UP000075881 UP000078492 UP000053825 UP000030765 UP000005203 UP000076407 UP000053105
UP000092443 UP000092462 UP000007798 UP000008792 UP000235965 UP000008711 UP000092460 UP000092445 UP000008744 UP000091820 UP000027135 UP000095301 UP000000304 UP000268350 UP000183832 UP000092444 UP000000803 UP000001292 UP000001819 UP000007801 UP000002282 UP000092553 UP000037069 UP000192221 UP000078200 UP000069940 UP000249989 UP000007819 UP000008820 UP000075886 UP000075901 UP000076408 UP000053097 UP000279307 UP000008237 UP000036403 UP000075884 UP000007266 UP000075880 UP000000673 UP000075883 UP000069272 UP000000311 UP000019118 UP000030742 UP000075920 UP000078542 UP000075902 UP000075882 UP000007062 UP000075885 UP000075903 UP000075840 UP000075881 UP000078492 UP000053825 UP000030765 UP000005203 UP000076407 UP000053105
PRIDE
Interpro
IPR029063
SAM-dependent_MTases
+ More
IPR001737 KsgA/Erm
IPR016861 TFB2M
IPR020598 rRNA_Ade_methylase_Trfase_N
IPR011037 Pyrv_Knase-like_insert_dom_sf
IPR036918 Pyrv_Knase_C_sf
IPR015806 Pyrv_Knase_insert_dom_sf
IPR015813 Pyrv/PenolPyrv_Kinase-like_dom
IPR040442 Pyrv_Kinase-like_dom_sf
IPR015793 Pyrv_Knase_brl
IPR001697 Pyr_Knase
IPR015795 Pyrv_Knase_C
IPR018209 Pyrv_Knase_AS
IPR031993 DUF4789
IPR035716 Cortactin_SH3
IPR003134 Hs1_Cortactin
IPR001452 SH3_domain
IPR036028 SH3-like_dom_sf
IPR001737 KsgA/Erm
IPR016861 TFB2M
IPR020598 rRNA_Ade_methylase_Trfase_N
IPR011037 Pyrv_Knase-like_insert_dom_sf
IPR036918 Pyrv_Knase_C_sf
IPR015806 Pyrv_Knase_insert_dom_sf
IPR015813 Pyrv/PenolPyrv_Kinase-like_dom
IPR040442 Pyrv_Kinase-like_dom_sf
IPR015793 Pyrv_Knase_brl
IPR001697 Pyr_Knase
IPR015795 Pyrv_Knase_C
IPR018209 Pyrv_Knase_AS
IPR031993 DUF4789
IPR035716 Cortactin_SH3
IPR003134 Hs1_Cortactin
IPR001452 SH3_domain
IPR036028 SH3-like_dom_sf
SUPFAM
Gene 3D
ProteinModelPortal
A0A2H1WV65
A0A1E1WHZ5
A0A1E1W153
A0A194PMD7
A0A194RKZ2
A0A212FE22
+ More
A0A034V410 A0A0A1X1H7 A0A034V534 A0A0K8VXA9 B4JSL0 A0A1I8QD22 W8AS10 B4KCG3 A0A1A9XNN1 A0A1B0DNZ7 B4NFL8 A0A1L8E141 B4M688 A0A2J7QNZ3 A0A0K8VKI4 B3P4M6 A0A1B0BK93 A0A1A9ZQ04 B4GZ09 A0A1A9VZA8 A0A067QQU3 A0A1I8MMU9 B4QVV5 A0A3B0K392 A0A1J1HU80 A0A1B0FHF6 Q9VH38 B4HJA9 Q29B70 B3M1F0 A0A2J7QNZ6 B4PVI5 A0A1B3PDF8 A0A0M5J4W3 A0A0L0BKZ3 A0A1W4W0K1 A0A1A9VNU8 A0A1B3PDB3 A0A023EYN0 A0A1Q3F632 A0A182H1Q2 A0A0K8U2I3 A0A336MXX8 J9JY26 A0A1B6D0A2 A0A1B6E0S9 A0A2S2QM08 A0A1S4FWI2 Q16LP3 A0A224XS25 A0A182QP86 A0A182TAI1 A0A182Y3T3 A0A026W748 E2BCN1 A0A0J7NQY4 Q16F37 A0A2H8TWS3 A0A182NRT5 A0A139WEP0 A0A2M4AWE9 A0A2M4AW36 A0A182JG70 W5JWF5 A0A182MR20 A0A182FP48 E2AVH6 N6SSI5 A0A182VZM2 A0A1Y1KCK8 A0A195C9S0 A0A1B6IKN4 A0A182TXU5 A0A182LA32 Q7PY27 A0A182PSI6 A0A182UTL6 A0A182HLN8 A0A182JU81 A0A1B6L546 A0A151IXH4 A0A0L7R1J5 A0A2M4BSH0 A0A2R7W7F0 A0A2M4BTB2 A0A2M4CNV7 A0A293N6V4 A0A084VLC0 A0A088A0E7 A0A182WST4 A0A0M8ZUN4 A0A0A9W2A9
A0A034V410 A0A0A1X1H7 A0A034V534 A0A0K8VXA9 B4JSL0 A0A1I8QD22 W8AS10 B4KCG3 A0A1A9XNN1 A0A1B0DNZ7 B4NFL8 A0A1L8E141 B4M688 A0A2J7QNZ3 A0A0K8VKI4 B3P4M6 A0A1B0BK93 A0A1A9ZQ04 B4GZ09 A0A1A9VZA8 A0A067QQU3 A0A1I8MMU9 B4QVV5 A0A3B0K392 A0A1J1HU80 A0A1B0FHF6 Q9VH38 B4HJA9 Q29B70 B3M1F0 A0A2J7QNZ6 B4PVI5 A0A1B3PDF8 A0A0M5J4W3 A0A0L0BKZ3 A0A1W4W0K1 A0A1A9VNU8 A0A1B3PDB3 A0A023EYN0 A0A1Q3F632 A0A182H1Q2 A0A0K8U2I3 A0A336MXX8 J9JY26 A0A1B6D0A2 A0A1B6E0S9 A0A2S2QM08 A0A1S4FWI2 Q16LP3 A0A224XS25 A0A182QP86 A0A182TAI1 A0A182Y3T3 A0A026W748 E2BCN1 A0A0J7NQY4 Q16F37 A0A2H8TWS3 A0A182NRT5 A0A139WEP0 A0A2M4AWE9 A0A2M4AW36 A0A182JG70 W5JWF5 A0A182MR20 A0A182FP48 E2AVH6 N6SSI5 A0A182VZM2 A0A1Y1KCK8 A0A195C9S0 A0A1B6IKN4 A0A182TXU5 A0A182LA32 Q7PY27 A0A182PSI6 A0A182UTL6 A0A182HLN8 A0A182JU81 A0A1B6L546 A0A151IXH4 A0A0L7R1J5 A0A2M4BSH0 A0A2R7W7F0 A0A2M4BTB2 A0A2M4CNV7 A0A293N6V4 A0A084VLC0 A0A088A0E7 A0A182WST4 A0A0M8ZUN4 A0A0A9W2A9
PDB
6ERO
E-value=0.0127687,
Score=90
Ontologies
GO
PANTHER
Topology
Subcellular location
Mitochondrion
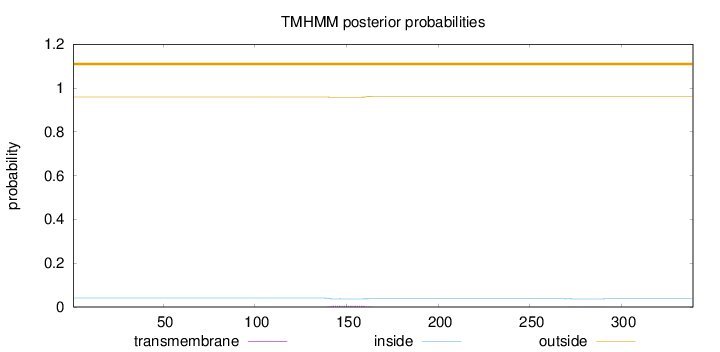
Length:
339
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.12087
Exp number, first 60 AAs:
0.00011
Total prob of N-in:
0.04133
outside
1 - 339
Population Genetic Test Statistics
Pi
231.278613
Theta
219.126892
Tajima's D
0.437823
CLR
62.56809
CSRT
0.498575071246438
Interpretation
Uncertain
