Gene
KWMTBOMO15152
Pre Gene Modal
BGIBMGA004930
Annotation
PREDICTED:_glucoside_xylosyltransferase_1_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.088 PlasmaMembrane Reliability : 1.533
Sequence
CDS
ATGAGAAATACGAAGCTATATTCCAATCATCAAAATAAGAGAATATTTAAATCGAAGTTGATGAAAAAAAATGATGAAGTTGTTTTATCATTTGTAGTATGTGACTCTAGGTTTAATGAGTCACTGAATTTGTTGAAGTCTGGACTTATTTTTACACAATCCTCCTTACATGTGGTTATATTTGCTGAAGATGATTTACACAAAAAGTTTAACAGTACATTGACAAAGTGGAAGATGTTGATTGCCAGACCATTCACTTTTGAACTTCATAAAATTAATTTTCCCGAAGCAAACAGAAAGGATTGGATGAACTTATTTAGTAAATGTGCTGCTCAAAGATTATTTATACCTGCATTGCTTCCTCATGTCGATGCTATGATATATGTGGATACAGACACCCTGTTTTTGGGTCCCATAGAAATTTTGTGGAATTACTTTACAAAATTTAAAGCTACAGAAATATCAGCTTTGGCTTTAGAAGATGAAAACCCAAATGTTTCATGGTACCCTAGATTTGCCAAACATCCATTTTATGGAAAATGCGGTTTGAACTCTGGTGTGATGCTAATGAATTTAACAAGGATGAGAGAATTTGGATGGATCGATTATATAACTCCTATCATGCTAAGGTGGAAATCCAATATACCGTGGGGCGACCAAGACATAATCAACATTATCTTTCACTATCACGAGAGGGCAGTCCATGAGTTGTCCTGCCGGTACAACTATCGTTCTGACCAGTGTATGTATGGTGACGCGTGCACAGATGCCACCTCCAACGGGATCTTCGTACTCCATGGCAGTCGCAGAGCATTCTATAGTGACAAGCAACCGGCGTTTCAAGCAGTGCACAGAGCTATAGAGGAGTTTGAAATTGGAGCAGATTTGTCGCCAAAAATGTTGCTCGTCAATATGGAGAAGTACTTGAACGAAACGCCCCCTTCGAATTGTGGAAACTTAAAAGATGCATTCCTGAAAATACCCACAGAGACCTTTAAAAAACATTACAACTTACCCGACAGATAG
Protein
MRNTKLYSNHQNKRIFKSKLMKKNDEVVLSFVVCDSRFNESLNLLKSGLIFTQSSLHVVIFAEDDLHKKFNSTLTKWKMLIARPFTFELHKINFPEANRKDWMNLFSKCAAQRLFIPALLPHVDAMIYVDTDTLFLGPIEILWNYFTKFKATEISALALEDENPNVSWYPRFAKHPFYGKCGLNSGVMLMNLTRMREFGWIDYITPIMLRWKSNIPWGDQDIINIIFHYHERAVHELSCRYNYRSDQCMYGDACTDATSNGIFVLHGSRRAFYSDKQPAFQAVHRAIEEFEIGADLSPKMLLVNMEKYLNETPPSNCGNLKDAFLKIPTETFKKHYNLPDR
Summary
Uniprot
H9J5Y9
A0A2H1WZD5
A0A2A4JMH9
A0A3S2NKR8
A0A212ETR5
A0A194RKZ7
+ More
A0A194PG51 A0A0L7KMS3 D6X1W8 A0A1B6CX52 J3JXR7 N6U2J6 A0A146L458 A0A0A9XB39 A0A0K8TEM9 A0A2A3EG76 A0A0L7QSW8 A0A087ZSQ1 A0A2J7R4M7 A0A1W4XLX4 A0A067RBN6 A0A1B6L8H4 A0A1Y1K506 A0A1B6HKJ2 A0A1B6EGF3 E2C236 A0A2S2QMF5 A0A026W2J0 E9IRV7 A0A0J7NJY2 A0A0C9R7Z6 A0A151J2Q5 A0A195FKV4 A0A195BF88 A0A158NUJ9 A0A151X6I6 F4WRD9 A0A2S2NXT1 A0A195C790 T1IDQ3 J9JP74 E2A383 A0A1B6F721 A0A0M9A1U1 A0A315VXX6 A0A3P9NX51 A0A3B5RF64 A0A0S7LD28 A0A154PD56 A0A3P9NX21 A0A1B0F073 M4AS78 A0A0S7LCG2 A0A232EG96 A0A087XHN3 A0A3B3VGR2 K7ISN2 A0A3P8UU56 A0A3B3VGQ3 A7S9E5 A0A3Q2DMV1 A0A3P8UU46 A0A2M4CQ93 A0A3Q2QCQ5 A0A060W0S4 A0A146V527 A0A146QTU1 W5UKV9 A0A3B4D2C1 W5JGS9 A0A3P8XXN2 A0A3P8S7Q0 A0A1S3MJ98 A0A3Q2NYA7 A0A3P8XXL9 A0A146ZL86 A0A3Q2CAN6 A0A3B5MVS0 A0A1S3MK96 A0A2D0SDV5 A0A2D0SDU0 A0A3P8S5E9 A0A3B4ZBY9 Q4RSH2 A0A146TJL9 A0A3Q1J8I7 A0A3B4CLY6 A0A2M4AUP9 A0A3Q1FAV7 A0A3P8ZTU9 A0A3Q2UT24 A0A3P8N7H5 A0A3B3BUA1 U5EUH3 A0A3Q0QUS2 G3NJ93 A0A0S7LCI5 A0A3B4F9I9 A0A3Q1J604 A0A182JPQ2
A0A194PG51 A0A0L7KMS3 D6X1W8 A0A1B6CX52 J3JXR7 N6U2J6 A0A146L458 A0A0A9XB39 A0A0K8TEM9 A0A2A3EG76 A0A0L7QSW8 A0A087ZSQ1 A0A2J7R4M7 A0A1W4XLX4 A0A067RBN6 A0A1B6L8H4 A0A1Y1K506 A0A1B6HKJ2 A0A1B6EGF3 E2C236 A0A2S2QMF5 A0A026W2J0 E9IRV7 A0A0J7NJY2 A0A0C9R7Z6 A0A151J2Q5 A0A195FKV4 A0A195BF88 A0A158NUJ9 A0A151X6I6 F4WRD9 A0A2S2NXT1 A0A195C790 T1IDQ3 J9JP74 E2A383 A0A1B6F721 A0A0M9A1U1 A0A315VXX6 A0A3P9NX51 A0A3B5RF64 A0A0S7LD28 A0A154PD56 A0A3P9NX21 A0A1B0F073 M4AS78 A0A0S7LCG2 A0A232EG96 A0A087XHN3 A0A3B3VGR2 K7ISN2 A0A3P8UU56 A0A3B3VGQ3 A7S9E5 A0A3Q2DMV1 A0A3P8UU46 A0A2M4CQ93 A0A3Q2QCQ5 A0A060W0S4 A0A146V527 A0A146QTU1 W5UKV9 A0A3B4D2C1 W5JGS9 A0A3P8XXN2 A0A3P8S7Q0 A0A1S3MJ98 A0A3Q2NYA7 A0A3P8XXL9 A0A146ZL86 A0A3Q2CAN6 A0A3B5MVS0 A0A1S3MK96 A0A2D0SDV5 A0A2D0SDU0 A0A3P8S5E9 A0A3B4ZBY9 Q4RSH2 A0A146TJL9 A0A3Q1J8I7 A0A3B4CLY6 A0A2M4AUP9 A0A3Q1FAV7 A0A3P8ZTU9 A0A3Q2UT24 A0A3P8N7H5 A0A3B3BUA1 U5EUH3 A0A3Q0QUS2 G3NJ93 A0A0S7LCI5 A0A3B4F9I9 A0A3Q1J604 A0A182JPQ2
Pubmed
EMBL
BABH01029961
BABH01029962
ODYU01012229
SOQ58423.1
NWSH01001030
PCG73029.1
+ More
RSAL01000053 RVE50086.1 AGBW02012531 OWR44878.1 KQ460045 KPJ18217.1 KQ459604 KPI92277.1 JTDY01008983 KOB64239.1 KQ971371 EFA10174.2 GEDC01019284 JAS18014.1 BT128038 AEE62999.1 APGK01044967 KB741028 KB631073 KB631074 ENN74846.1 ERL84072.1 ERL84081.1 GDHC01016204 GDHC01015474 JAQ02425.1 JAQ03155.1 GBHO01025662 GBHO01017435 JAG17942.1 JAG26169.1 GBRD01002216 JAG63605.1 KZ288266 PBC30156.1 KQ414756 KOC61649.1 NEVH01007405 PNF35775.1 KK852570 KDR21132.1 GEBQ01019965 JAT20012.1 GEZM01098287 JAV53927.1 GECU01032545 JAS75161.1 GEDC01000272 JAS37026.1 GL452067 EFN77987.1 GGMS01009660 MBY78863.1 KK107496 QOIP01000004 EZA49811.1 RLU24096.1 GL765283 EFZ16676.1 LBMM01004124 KMQ92790.1 GBYB01012514 GBYB01012515 JAG82281.1 JAG82282.1 KQ980368 KYN16332.1 KQ981522 KYN40624.1 KQ976504 KYM82857.1 ADTU01026586 KQ982481 KYQ55996.1 GL888285 EGI63210.1 GGMR01009371 MBY21990.1 KQ978143 KYM96712.1 ACPB03000197 ABLF02019095 GL436311 EFN72111.1 GECZ01023733 JAS46036.1 KQ435760 KOX75575.1 NHOQ01000831 PWA28469.1 GBYX01176107 JAO87194.1 KQ434868 KZC09188.1 AJVK01007413 GBYX01176113 JAO87190.1 NNAY01004813 OXU17380.1 AYCK01002244 AYCK01002245 AAZX01011284 DS469603 EDO39657.1 GGFL01003334 MBW67512.1 FR904298 CDQ58175.1 GCES01074194 JAR12129.1 GCES01126823 JAQ59499.1 JT418339 AHH42702.1 ADMH02001231 ETN63567.1 GCES01019212 JAR67111.1 CAAE01015000 CAG08660.1 GCES01093515 JAQ92807.1 GGFK01011182 MBW44503.1 GANO01003880 JAB55991.1 GBYX01176109 JAO87192.1
RSAL01000053 RVE50086.1 AGBW02012531 OWR44878.1 KQ460045 KPJ18217.1 KQ459604 KPI92277.1 JTDY01008983 KOB64239.1 KQ971371 EFA10174.2 GEDC01019284 JAS18014.1 BT128038 AEE62999.1 APGK01044967 KB741028 KB631073 KB631074 ENN74846.1 ERL84072.1 ERL84081.1 GDHC01016204 GDHC01015474 JAQ02425.1 JAQ03155.1 GBHO01025662 GBHO01017435 JAG17942.1 JAG26169.1 GBRD01002216 JAG63605.1 KZ288266 PBC30156.1 KQ414756 KOC61649.1 NEVH01007405 PNF35775.1 KK852570 KDR21132.1 GEBQ01019965 JAT20012.1 GEZM01098287 JAV53927.1 GECU01032545 JAS75161.1 GEDC01000272 JAS37026.1 GL452067 EFN77987.1 GGMS01009660 MBY78863.1 KK107496 QOIP01000004 EZA49811.1 RLU24096.1 GL765283 EFZ16676.1 LBMM01004124 KMQ92790.1 GBYB01012514 GBYB01012515 JAG82281.1 JAG82282.1 KQ980368 KYN16332.1 KQ981522 KYN40624.1 KQ976504 KYM82857.1 ADTU01026586 KQ982481 KYQ55996.1 GL888285 EGI63210.1 GGMR01009371 MBY21990.1 KQ978143 KYM96712.1 ACPB03000197 ABLF02019095 GL436311 EFN72111.1 GECZ01023733 JAS46036.1 KQ435760 KOX75575.1 NHOQ01000831 PWA28469.1 GBYX01176107 JAO87194.1 KQ434868 KZC09188.1 AJVK01007413 GBYX01176113 JAO87190.1 NNAY01004813 OXU17380.1 AYCK01002244 AYCK01002245 AAZX01011284 DS469603 EDO39657.1 GGFL01003334 MBW67512.1 FR904298 CDQ58175.1 GCES01074194 JAR12129.1 GCES01126823 JAQ59499.1 JT418339 AHH42702.1 ADMH02001231 ETN63567.1 GCES01019212 JAR67111.1 CAAE01015000 CAG08660.1 GCES01093515 JAQ92807.1 GGFK01011182 MBW44503.1 GANO01003880 JAB55991.1 GBYX01176109 JAO87192.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000007151
UP000053240
UP000053268
+ More
UP000037510 UP000007266 UP000019118 UP000030742 UP000242457 UP000053825 UP000005203 UP000235965 UP000192223 UP000027135 UP000008237 UP000053097 UP000279307 UP000036403 UP000078492 UP000078541 UP000078540 UP000005205 UP000075809 UP000007755 UP000078542 UP000015103 UP000007819 UP000000311 UP000053105 UP000242638 UP000002852 UP000076502 UP000092462 UP000215335 UP000028760 UP000261500 UP000002358 UP000265120 UP000001593 UP000265020 UP000265000 UP000193380 UP000221080 UP000261440 UP000000673 UP000265140 UP000265080 UP000087266 UP000261380 UP000261400 UP000265040 UP000257200 UP000264840 UP000265100 UP000261560 UP000261340 UP000007635 UP000261460 UP000075881
UP000037510 UP000007266 UP000019118 UP000030742 UP000242457 UP000053825 UP000005203 UP000235965 UP000192223 UP000027135 UP000008237 UP000053097 UP000279307 UP000036403 UP000078492 UP000078541 UP000078540 UP000005205 UP000075809 UP000007755 UP000078542 UP000015103 UP000007819 UP000000311 UP000053105 UP000242638 UP000002852 UP000076502 UP000092462 UP000215335 UP000028760 UP000261500 UP000002358 UP000265120 UP000001593 UP000265020 UP000265000 UP000193380 UP000221080 UP000261440 UP000000673 UP000265140 UP000265080 UP000087266 UP000261380 UP000261400 UP000265040 UP000257200 UP000264840 UP000265100 UP000261560 UP000261340 UP000007635 UP000261460 UP000075881
PRIDE
Pfam
PF01501 Glyco_transf_8
SUPFAM
SSF53448
SSF53448
Gene 3D
ProteinModelPortal
H9J5Y9
A0A2H1WZD5
A0A2A4JMH9
A0A3S2NKR8
A0A212ETR5
A0A194RKZ7
+ More
A0A194PG51 A0A0L7KMS3 D6X1W8 A0A1B6CX52 J3JXR7 N6U2J6 A0A146L458 A0A0A9XB39 A0A0K8TEM9 A0A2A3EG76 A0A0L7QSW8 A0A087ZSQ1 A0A2J7R4M7 A0A1W4XLX4 A0A067RBN6 A0A1B6L8H4 A0A1Y1K506 A0A1B6HKJ2 A0A1B6EGF3 E2C236 A0A2S2QMF5 A0A026W2J0 E9IRV7 A0A0J7NJY2 A0A0C9R7Z6 A0A151J2Q5 A0A195FKV4 A0A195BF88 A0A158NUJ9 A0A151X6I6 F4WRD9 A0A2S2NXT1 A0A195C790 T1IDQ3 J9JP74 E2A383 A0A1B6F721 A0A0M9A1U1 A0A315VXX6 A0A3P9NX51 A0A3B5RF64 A0A0S7LD28 A0A154PD56 A0A3P9NX21 A0A1B0F073 M4AS78 A0A0S7LCG2 A0A232EG96 A0A087XHN3 A0A3B3VGR2 K7ISN2 A0A3P8UU56 A0A3B3VGQ3 A7S9E5 A0A3Q2DMV1 A0A3P8UU46 A0A2M4CQ93 A0A3Q2QCQ5 A0A060W0S4 A0A146V527 A0A146QTU1 W5UKV9 A0A3B4D2C1 W5JGS9 A0A3P8XXN2 A0A3P8S7Q0 A0A1S3MJ98 A0A3Q2NYA7 A0A3P8XXL9 A0A146ZL86 A0A3Q2CAN6 A0A3B5MVS0 A0A1S3MK96 A0A2D0SDV5 A0A2D0SDU0 A0A3P8S5E9 A0A3B4ZBY9 Q4RSH2 A0A146TJL9 A0A3Q1J8I7 A0A3B4CLY6 A0A2M4AUP9 A0A3Q1FAV7 A0A3P8ZTU9 A0A3Q2UT24 A0A3P8N7H5 A0A3B3BUA1 U5EUH3 A0A3Q0QUS2 G3NJ93 A0A0S7LCI5 A0A3B4F9I9 A0A3Q1J604 A0A182JPQ2
A0A194PG51 A0A0L7KMS3 D6X1W8 A0A1B6CX52 J3JXR7 N6U2J6 A0A146L458 A0A0A9XB39 A0A0K8TEM9 A0A2A3EG76 A0A0L7QSW8 A0A087ZSQ1 A0A2J7R4M7 A0A1W4XLX4 A0A067RBN6 A0A1B6L8H4 A0A1Y1K506 A0A1B6HKJ2 A0A1B6EGF3 E2C236 A0A2S2QMF5 A0A026W2J0 E9IRV7 A0A0J7NJY2 A0A0C9R7Z6 A0A151J2Q5 A0A195FKV4 A0A195BF88 A0A158NUJ9 A0A151X6I6 F4WRD9 A0A2S2NXT1 A0A195C790 T1IDQ3 J9JP74 E2A383 A0A1B6F721 A0A0M9A1U1 A0A315VXX6 A0A3P9NX51 A0A3B5RF64 A0A0S7LD28 A0A154PD56 A0A3P9NX21 A0A1B0F073 M4AS78 A0A0S7LCG2 A0A232EG96 A0A087XHN3 A0A3B3VGR2 K7ISN2 A0A3P8UU56 A0A3B3VGQ3 A7S9E5 A0A3Q2DMV1 A0A3P8UU46 A0A2M4CQ93 A0A3Q2QCQ5 A0A060W0S4 A0A146V527 A0A146QTU1 W5UKV9 A0A3B4D2C1 W5JGS9 A0A3P8XXN2 A0A3P8S7Q0 A0A1S3MJ98 A0A3Q2NYA7 A0A3P8XXL9 A0A146ZL86 A0A3Q2CAN6 A0A3B5MVS0 A0A1S3MK96 A0A2D0SDV5 A0A2D0SDU0 A0A3P8S5E9 A0A3B4ZBY9 Q4RSH2 A0A146TJL9 A0A3Q1J8I7 A0A3B4CLY6 A0A2M4AUP9 A0A3Q1FAV7 A0A3P8ZTU9 A0A3Q2UT24 A0A3P8N7H5 A0A3B3BUA1 U5EUH3 A0A3Q0QUS2 G3NJ93 A0A0S7LCI5 A0A3B4F9I9 A0A3Q1J604 A0A182JPQ2
PDB
4WNH
E-value=0.00374634,
Score=94
Ontologies
GO
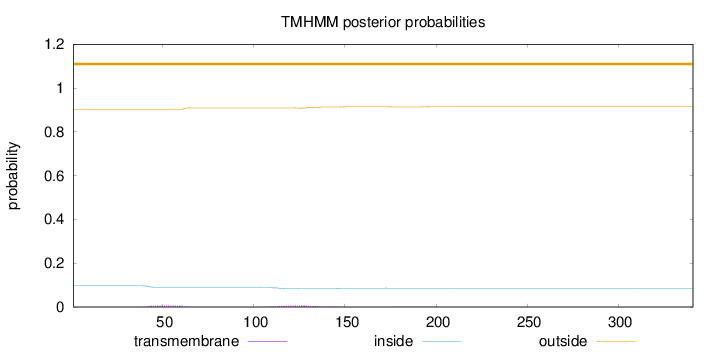
Topology
Length:
341
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.30655
Exp number, first 60 AAs:
0.12354
Total prob of N-in:
0.09714
outside
1 - 341
Population Genetic Test Statistics
Pi
214.59738
Theta
188.414814
Tajima's D
0.470649
CLR
0.101134
CSRT
0.509674516274186
Interpretation
Possibly Positive selection
