Gene
KWMTBOMO15151
Pre Gene Modal
BGIBMGA004985
Annotation
DNA-directed_RNA_polymerase_III_subunit_RPC6_[Operophtera_brumata]
Full name
DNA-directed RNA polymerase III subunit RPC6
Alternative Name
DNA-directed RNA polymerase III subunit F
RNA polymerase III 39 kDa subunit
RNA polymerase III 39 kDa subunit
Location in the cell
Cytoplasmic Reliability : 1.488 Nuclear Reliability : 1.805
Sequence
CDS
ATGAGTTCCGAAGATACAAAAGAGAAAATACTAAATCTTGCAAGAAAAAGCTCGAAAGGAATTACAGACAAGGATATAATTGCTGAAATACCTGGCATATCATCAGTAGAACTAGTAACTGCTTTAAATGATCTGCTGAAACAAGAACGTTTTGATTTATTTAATCAAAACGGATCTTTAATTTATAAGTTGAAAACTCAAACAAATAAGCACGCAATCAAAGGAGCAGATAATGAAGAAAAGGTAGTCTATAATCTGATTGAAGAAGCAGGAAATAAAGGAATTTGGATTAGAGATATTAGGATCCGTTCGAATCTTGCAAACACTCAACTGACTAAAGTCCTCAAAAATTTGGAAAGTAAAAAAGTGATAAAAGCTGTGAAATGTGTTAATGCCTCTAAAAAGAAGGTTTATATGCTTTACAACTTGGAACCAGATAGATCATTGACAGGGGGAGCTTGGTACCAAGACCAAGACTTTGAATCCGAGTTTGTAGATGTCCTCAATGTACAATGCCACAGGTTCTTGAGTCAAAGAGCTGATAATATGAGAAACAATCCTAGGGGACCTGTTTTTGGACGAACACAAGCTTATGCTACTGCTACTGAAGTCCAAAAATATATTACTGATCTGGGAATTAGTAAAGTTGTATTAGATGTTGATGATGTAACAACAATTTTAAACACCCTAGTGTATGATGGGAAGGTAGAAAAAAGTGTGTTTCCCGATGGAAGTAATGTATATAGAGCTATAGAAACTCTTATACCATCTTCAGGCCTTGTTCAAGTGCCATGTGGAATATGCCCCTTGATTCAAAGATGTAATTCCTCAGGACTAATTACTCCACAAGTTTGTAATTATATGACTGAATGGTTGGAATAA
Protein
MSSEDTKEKILNLARKSSKGITDKDIIAEIPGISSVELVTALNDLLKQERFDLFNQNGSLIYKLKTQTNKHAIKGADNEEKVVYNLIEEAGNKGIWIRDIRIRSNLANTQLTKVLKNLESKKVIKAVKCVNASKKKVYMLYNLEPDRSLTGGAWYQDQDFESEFVDVLNVQCHRFLSQRADNMRNNPRGPVFGRTQAYATATEVQKYITDLGISKVVLDVDDVTTILNTLVYDGKVEKSVFPDGSNVYRAIETLIPSSGLVQVPCGICPLIQRCNSSGLITPQVCNYMTEWLE
Summary
Description
DNA-dependent RNA polymerase catalyzes the transcription of DNA into RNA using the four ribonucleoside triphosphates as substrates. Specific peripheric component of RNA polymerase III which synthesizes small RNAs, such as 5S rRNA and tRNAs.
DNA-dependent RNA polymerase catalyzes the transcription of DNA into RNA using the four ribonucleoside triphosphates as substrates. Specific peripheric component of RNA polymerase III which synthesizes small RNAs, such as 5S rRNA and tRNAs. May direct RNA Pol III binding to the TFIIIB-DNA complex. Plays a key role in sensing and limiting infection by intracellular bacteria and DNA viruses. Acts as nuclear and cytosolic DNA sensor involved in innate immune response. Can sense non-self dsDNA that serves as template for transcription into dsRNA. The non-self RNA polymerase III transcripts induce type I interferon and NF- Kappa-B through the RIG-I pathway (By similarity). Preferentially binds double-stranded DNA (dsDNA) (By similarity).
DNA-dependent RNA polymerase catalyzes the transcription of DNA into RNA using the four ribonucleoside triphosphates as substrates. Specific peripheric component of RNA polymerase III which synthesizes small RNAs, such as 5S rRNA and tRNAs. May direct RNA Pol III binding to the TFIIIB-DNA complex. Plays a key role in sensing and limiting infection by intracellular bacteria and DNA viruses. Acts as nuclear and cytosolic DNA sensor involved in innate immune response. Can sense non-self dsDNA that serves as template for transcription into dsRNA. The non-self RNA polymerase III transcripts, such as Epstein-Barr virus-encoded RNAs (EBERs) induce type I interferon and NF- Kappa-B through the RIG-I pathway. Preferentially binds double-stranded DNA (dsDNA) (PubMed:21358628).
DNA-dependent RNA polymerase catalyzes the transcription of DNA into RNA using the four ribonucleoside triphosphates as substrates. Specific peripheric component of RNA polymerase III which synthesizes small RNAs, such as 5S rRNA and tRNAs. May direct RNA Pol III binding to the TFIIIB-DNA complex. Plays a key role in sensing and limiting infection by intracellular bacteria and DNA viruses. Acts as nuclear and cytosolic DNA sensor involved in innate immune response. Can sense non-self dsDNA that serves as template for transcription into dsRNA. The non-self RNA polymerase III transcripts induce type I interferon and NF- Kappa-B through the RIG-I pathway (By similarity). Preferentially binds double-stranded DNA (dsDNA) (By similarity).
DNA-dependent RNA polymerase catalyzes the transcription of DNA into RNA using the four ribonucleoside triphosphates as substrates. Specific peripheric component of RNA polymerase III which synthesizes small RNAs, such as 5S rRNA and tRNAs. May direct RNA Pol III binding to the TFIIIB-DNA complex. Plays a key role in sensing and limiting infection by intracellular bacteria and DNA viruses. Acts as nuclear and cytosolic DNA sensor involved in innate immune response. Can sense non-self dsDNA that serves as template for transcription into dsRNA. The non-self RNA polymerase III transcripts, such as Epstein-Barr virus-encoded RNAs (EBERs) induce type I interferon and NF- Kappa-B through the RIG-I pathway. Preferentially binds double-stranded DNA (dsDNA) (PubMed:21358628).
Subunit
Component of the RNA polymerase III (Pol III) complex consisting of 17 subunits. RPC3/POLR3C, RPC6/POLR3F and RPC7/POLR3G form a Pol III subcomplex (By similarity). Directly interacts with POLR3C (By similarity). Interacts with TBP and TFIIIB90 and GTF3C4 (By similarity). Interacts with MAF1 (By similarity).
Component of the RNA polymerase III (Pol III) complex consisting of 17 subunits. RPC3/POLR3C, RPC6/POLR3F and RPC7/POLR3G form a Pol III subcomplex (By similarity). Directly interacts with POLR3C (PubMed:21358628). Interacts with TBP and TFIIIB90 and GTF3C4 (By similarity) (PubMed:10523658). Interacts with MAF1 (PubMed:18377933).
Component of the RNA polymerase III (Pol III) complex consisting of 17 subunits. RPC3/POLR3C, RPC6/POLR3F and RPC7/POLR3G form a Pol III subcomplex (By similarity). Directly interacts with POLR3C (PubMed:21358628). Interacts with TBP and TFIIIB90 and GTF3C4 (By similarity) (PubMed:10523658). Interacts with MAF1 (PubMed:18377933).
Similarity
Belongs to the eukaryotic RPC34/RPC39 RNA polymerase subunit family.
Keywords
Acetylation
Antiviral defense
Complete proteome
DNA-directed RNA polymerase
Immunity
Innate immunity
Isopeptide bond
Nucleus
Reference proteome
Transcription
Ubl conjugation
3D-structure
DNA-binding
Feature
chain DNA-directed RNA polymerase III subunit RPC6
Uniprot
H9J644
A0A2A4JMS8
A0A2H1X1B9
A0A0L7K528
A0A3S2LBL6
S4PFD3
+ More
A0A194PMD1 A0A194RLC3 I4DPJ5 A0A212ETQ9 A0A067R809 A0A2J7R275 A0A2J7R283 A0A0N0U431 A0A232ETH4 K7J1D6 A0A2A3EQF8 E9IUD9 A0A0L7R1J8 A0A087ZZS0 A0A026WU45 A0A154P4N9 A0A3L8D8Z5 A0A151WH64 E2AN67 A0A3L8D8H1 E2BG67 E0W056 A0A1L8DH24 A0A158NA11 A0A1Y1KIJ9 U5ELI2 A0A1I8P898 J3JTQ9 N6TQ48 A0A1B6LXI3 F4WC50 A0A1I8MSH0 A0A1B6DXE9 A0A195FQZ6 W8AF96 A0A1B6HCU6 A0A084W9B5 A0A182GJQ4 A0A023EP59 A0A182Q5K6 A0A0K8UN47 A0A151J0N8 A0A0L0BMZ5 D6WU15 A0A034WW18 S4RLM9 Q17EJ2 B0XD81 A0A1W4X6H6 A0A2R5L7G7 A0A182K081 C3XZ17 A0A1Q3F687 Q7PZL8 A0A195CUS1 A0A0L8FIU6 A0A182YBN2 A0A2K6EI85 A0A182L5B3 A0A182LSY9 G1T8N5 A0A182NZY6 A0A182W232 H0UYH0 K9IJ69 A0A2K6C5Y0 A0A2K6K0B3 A0A2K5XNF2 A0A0D9RGH0 A0A096NVN9 A0A2K6QUU0 A0A2K5KZY0 G7PH18 G7N320 A0A131XXX8 A0A1U7TY73 Q2T9S3 G3R7C7 G1RFC5 F6RMB3 A0A2R9BKK2 H2P144 H2QK06 Q9H1D9 H3ALW7 A0A147BQU5 D2HSX3 L8J1H4 W5NQ69 F1MYX7 E2RG78 A0A3Q7SCX0 A8QKB1 Q921X6
A0A194PMD1 A0A194RLC3 I4DPJ5 A0A212ETQ9 A0A067R809 A0A2J7R275 A0A2J7R283 A0A0N0U431 A0A232ETH4 K7J1D6 A0A2A3EQF8 E9IUD9 A0A0L7R1J8 A0A087ZZS0 A0A026WU45 A0A154P4N9 A0A3L8D8Z5 A0A151WH64 E2AN67 A0A3L8D8H1 E2BG67 E0W056 A0A1L8DH24 A0A158NA11 A0A1Y1KIJ9 U5ELI2 A0A1I8P898 J3JTQ9 N6TQ48 A0A1B6LXI3 F4WC50 A0A1I8MSH0 A0A1B6DXE9 A0A195FQZ6 W8AF96 A0A1B6HCU6 A0A084W9B5 A0A182GJQ4 A0A023EP59 A0A182Q5K6 A0A0K8UN47 A0A151J0N8 A0A0L0BMZ5 D6WU15 A0A034WW18 S4RLM9 Q17EJ2 B0XD81 A0A1W4X6H6 A0A2R5L7G7 A0A182K081 C3XZ17 A0A1Q3F687 Q7PZL8 A0A195CUS1 A0A0L8FIU6 A0A182YBN2 A0A2K6EI85 A0A182L5B3 A0A182LSY9 G1T8N5 A0A182NZY6 A0A182W232 H0UYH0 K9IJ69 A0A2K6C5Y0 A0A2K6K0B3 A0A2K5XNF2 A0A0D9RGH0 A0A096NVN9 A0A2K6QUU0 A0A2K5KZY0 G7PH18 G7N320 A0A131XXX8 A0A1U7TY73 Q2T9S3 G3R7C7 G1RFC5 F6RMB3 A0A2R9BKK2 H2P144 H2QK06 Q9H1D9 H3ALW7 A0A147BQU5 D2HSX3 L8J1H4 W5NQ69 F1MYX7 E2RG78 A0A3Q7SCX0 A8QKB1 Q921X6
Pubmed
19121390
26227816
23622113
26354079
22651552
22118469
+ More
24845553 28648823 20075255 21282665 24508170 30249741 20798317 20566863 21347285 28004739 22516182 23537049 21719571 25315136 24495485 24438588 26483478 24945155 26108605 18362917 19820115 25348373 17510324 18563158 12364791 14747013 17210077 25244985 20966253 21993624 25362486 22002653 25319552 22398555 19892987 22722832 16136131 9171375 14702039 11780052 15489334 10523658 12391170 18377933 21269460 21358628 22814378 25218447 25772364 28112733 19631370 19609254 9215903 29652888 20010809 22751099 20809919 19393038 16341006 17536020 16141072 21183079
24845553 28648823 20075255 21282665 24508170 30249741 20798317 20566863 21347285 28004739 22516182 23537049 21719571 25315136 24495485 24438588 26483478 24945155 26108605 18362917 19820115 25348373 17510324 18563158 12364791 14747013 17210077 25244985 20966253 21993624 25362486 22002653 25319552 22398555 19892987 22722832 16136131 9171375 14702039 11780052 15489334 10523658 12391170 18377933 21269460 21358628 22814378 25218447 25772364 28112733 19631370 19609254 9215903 29652888 20010809 22751099 20809919 19393038 16341006 17536020 16141072 21183079
EMBL
BABH01029964
NWSH01001030
PCG73028.1
ODYU01012229
SOQ58424.1
JTDY01011012
+ More
KOB57702.1 RSAL01000053 RVE50084.1 GAIX01004067 JAA88493.1 KQ459604 KPI92275.1 KQ460045 KPJ18219.1 AK403638 BAM19835.1 AGBW02012531 OWR44880.1 KK852643 KDR19576.1 NEVH01008200 PNF34937.1 PNF34936.1 KQ435863 KOX70382.1 NNAY01002276 OXU21651.1 KZ288194 PBC33920.1 GL765967 EFZ15814.1 KQ414667 KOC64719.1 KK107105 EZA59552.1 KQ434809 KZC06178.1 QOIP01000011 RLU16583.1 KQ983135 KYQ47164.1 GL441088 EFN65131.1 RLU16581.1 GL448115 EFN85335.1 DS235857 EEB19012.1 GFDF01008440 JAV05644.1 ADTU01009933 GEZM01085246 GEZM01085245 JAV60050.1 GANO01001395 JAB58476.1 BT126616 AEE61580.1 APGK01059162 KB741292 KB632218 ENN70441.1 ERL90176.1 GEBQ01011624 JAT28353.1 GL888070 EGI68030.1 GEDC01021554 GEDC01006946 JAS15744.1 JAS30352.1 KQ981305 KYN42893.1 GAMC01019656 GAMC01019653 GAMC01019645 JAB86910.1 GECU01035200 JAS72506.1 ATLV01021669 KE525321 KFB46809.1 JXUM01068660 KQ562512 KXJ75740.1 GAPW01002838 JAC10760.1 AXCN02001144 GDHF01031334 GDHF01024185 GDHF01023378 GDHF01016551 GDHF01005231 JAI20980.1 JAI28129.1 JAI28936.1 JAI35763.1 JAI47083.1 KQ980629 KYN15022.1 JRES01001623 KNC21391.1 KQ971352 EFA07655.1 GAKP01000395 JAC58557.1 CH477282 EAT44887.1 DS232745 EDS45335.1 GGLE01001251 MBY05377.1 GG666475 EEN66581.1 GFDL01011961 JAV23084.1 AAAB01008986 EAA00356.3 KQ977276 KYN04431.1 KQ431108 KOF63341.1 AXCM01000173 AAGW02052459 AAKN02012609 GABZ01007063 JAA46462.1 AQIB01145272 AQIB01145273 AHZZ02000689 AQIA01008037 AQIA01008038 CM001285 EHH65574.1 JU320037 JU471482 CM001262 AFE63793.1 AFH28286.1 EHH19952.1 GEFM01005240 JAP70556.1 BC111292 CABD030115794 ADFV01042570 AJFE02032906 ABGA01338643 NDHI03003422 PNJ55536.1 AACZ04064906 GABC01008529 GABF01003348 GABD01007628 GABE01010756 NBAG03000245 JAA02809.1 JAA18797.1 JAA25472.1 JAA33983.1 PNI62693.1 U93869 AK290892 AL121893 CH471133 BC012588 AFYH01043398 AFYH01043399 AFYH01043400 AFYH01043401 AFYH01043402 AFYH01043403 GEGO01002532 JAR92872.1 GL193309 EFB28253.1 JH880468 ELR61252.1 AMGL01024942 AAEX03013766 EF068179 ABN72532.1 AK036166 AK076538 AK144679 BC009159
KOB57702.1 RSAL01000053 RVE50084.1 GAIX01004067 JAA88493.1 KQ459604 KPI92275.1 KQ460045 KPJ18219.1 AK403638 BAM19835.1 AGBW02012531 OWR44880.1 KK852643 KDR19576.1 NEVH01008200 PNF34937.1 PNF34936.1 KQ435863 KOX70382.1 NNAY01002276 OXU21651.1 KZ288194 PBC33920.1 GL765967 EFZ15814.1 KQ414667 KOC64719.1 KK107105 EZA59552.1 KQ434809 KZC06178.1 QOIP01000011 RLU16583.1 KQ983135 KYQ47164.1 GL441088 EFN65131.1 RLU16581.1 GL448115 EFN85335.1 DS235857 EEB19012.1 GFDF01008440 JAV05644.1 ADTU01009933 GEZM01085246 GEZM01085245 JAV60050.1 GANO01001395 JAB58476.1 BT126616 AEE61580.1 APGK01059162 KB741292 KB632218 ENN70441.1 ERL90176.1 GEBQ01011624 JAT28353.1 GL888070 EGI68030.1 GEDC01021554 GEDC01006946 JAS15744.1 JAS30352.1 KQ981305 KYN42893.1 GAMC01019656 GAMC01019653 GAMC01019645 JAB86910.1 GECU01035200 JAS72506.1 ATLV01021669 KE525321 KFB46809.1 JXUM01068660 KQ562512 KXJ75740.1 GAPW01002838 JAC10760.1 AXCN02001144 GDHF01031334 GDHF01024185 GDHF01023378 GDHF01016551 GDHF01005231 JAI20980.1 JAI28129.1 JAI28936.1 JAI35763.1 JAI47083.1 KQ980629 KYN15022.1 JRES01001623 KNC21391.1 KQ971352 EFA07655.1 GAKP01000395 JAC58557.1 CH477282 EAT44887.1 DS232745 EDS45335.1 GGLE01001251 MBY05377.1 GG666475 EEN66581.1 GFDL01011961 JAV23084.1 AAAB01008986 EAA00356.3 KQ977276 KYN04431.1 KQ431108 KOF63341.1 AXCM01000173 AAGW02052459 AAKN02012609 GABZ01007063 JAA46462.1 AQIB01145272 AQIB01145273 AHZZ02000689 AQIA01008037 AQIA01008038 CM001285 EHH65574.1 JU320037 JU471482 CM001262 AFE63793.1 AFH28286.1 EHH19952.1 GEFM01005240 JAP70556.1 BC111292 CABD030115794 ADFV01042570 AJFE02032906 ABGA01338643 NDHI03003422 PNJ55536.1 AACZ04064906 GABC01008529 GABF01003348 GABD01007628 GABE01010756 NBAG03000245 JAA02809.1 JAA18797.1 JAA25472.1 JAA33983.1 PNI62693.1 U93869 AK290892 AL121893 CH471133 BC012588 AFYH01043398 AFYH01043399 AFYH01043400 AFYH01043401 AFYH01043402 AFYH01043403 GEGO01002532 JAR92872.1 GL193309 EFB28253.1 JH880468 ELR61252.1 AMGL01024942 AAEX03013766 EF068179 ABN72532.1 AK036166 AK076538 AK144679 BC009159
Proteomes
UP000005204
UP000218220
UP000037510
UP000283053
UP000053268
UP000053240
+ More
UP000007151 UP000027135 UP000235965 UP000053105 UP000215335 UP000002358 UP000242457 UP000053825 UP000005203 UP000053097 UP000076502 UP000279307 UP000075809 UP000000311 UP000008237 UP000009046 UP000005205 UP000095300 UP000019118 UP000030742 UP000007755 UP000095301 UP000078541 UP000030765 UP000069940 UP000249989 UP000075886 UP000078492 UP000037069 UP000007266 UP000245300 UP000008820 UP000002320 UP000192223 UP000075881 UP000001554 UP000007062 UP000078542 UP000053454 UP000076408 UP000233160 UP000075882 UP000075883 UP000001811 UP000075885 UP000075920 UP000005447 UP000233120 UP000233180 UP000233140 UP000029965 UP000028761 UP000233200 UP000233060 UP000009130 UP000233100 UP000189704 UP000009136 UP000001519 UP000001073 UP000002281 UP000240080 UP000001595 UP000002277 UP000005640 UP000008672 UP000002356 UP000002254 UP000286640 UP000000589
UP000007151 UP000027135 UP000235965 UP000053105 UP000215335 UP000002358 UP000242457 UP000053825 UP000005203 UP000053097 UP000076502 UP000279307 UP000075809 UP000000311 UP000008237 UP000009046 UP000005205 UP000095300 UP000019118 UP000030742 UP000007755 UP000095301 UP000078541 UP000030765 UP000069940 UP000249989 UP000075886 UP000078492 UP000037069 UP000007266 UP000245300 UP000008820 UP000002320 UP000192223 UP000075881 UP000001554 UP000007062 UP000078542 UP000053454 UP000076408 UP000233160 UP000075882 UP000075883 UP000001811 UP000075885 UP000075920 UP000005447 UP000233120 UP000233180 UP000233140 UP000029965 UP000028761 UP000233200 UP000233060 UP000009130 UP000233100 UP000189704 UP000009136 UP000001519 UP000001073 UP000002281 UP000240080 UP000001595 UP000002277 UP000005640 UP000008672 UP000002356 UP000002254 UP000286640 UP000000589
Pfam
PF05158 RNA_pol_Rpc34
Interpro
SUPFAM
SSF46785
SSF46785
Gene 3D
ProteinModelPortal
H9J644
A0A2A4JMS8
A0A2H1X1B9
A0A0L7K528
A0A3S2LBL6
S4PFD3
+ More
A0A194PMD1 A0A194RLC3 I4DPJ5 A0A212ETQ9 A0A067R809 A0A2J7R275 A0A2J7R283 A0A0N0U431 A0A232ETH4 K7J1D6 A0A2A3EQF8 E9IUD9 A0A0L7R1J8 A0A087ZZS0 A0A026WU45 A0A154P4N9 A0A3L8D8Z5 A0A151WH64 E2AN67 A0A3L8D8H1 E2BG67 E0W056 A0A1L8DH24 A0A158NA11 A0A1Y1KIJ9 U5ELI2 A0A1I8P898 J3JTQ9 N6TQ48 A0A1B6LXI3 F4WC50 A0A1I8MSH0 A0A1B6DXE9 A0A195FQZ6 W8AF96 A0A1B6HCU6 A0A084W9B5 A0A182GJQ4 A0A023EP59 A0A182Q5K6 A0A0K8UN47 A0A151J0N8 A0A0L0BMZ5 D6WU15 A0A034WW18 S4RLM9 Q17EJ2 B0XD81 A0A1W4X6H6 A0A2R5L7G7 A0A182K081 C3XZ17 A0A1Q3F687 Q7PZL8 A0A195CUS1 A0A0L8FIU6 A0A182YBN2 A0A2K6EI85 A0A182L5B3 A0A182LSY9 G1T8N5 A0A182NZY6 A0A182W232 H0UYH0 K9IJ69 A0A2K6C5Y0 A0A2K6K0B3 A0A2K5XNF2 A0A0D9RGH0 A0A096NVN9 A0A2K6QUU0 A0A2K5KZY0 G7PH18 G7N320 A0A131XXX8 A0A1U7TY73 Q2T9S3 G3R7C7 G1RFC5 F6RMB3 A0A2R9BKK2 H2P144 H2QK06 Q9H1D9 H3ALW7 A0A147BQU5 D2HSX3 L8J1H4 W5NQ69 F1MYX7 E2RG78 A0A3Q7SCX0 A8QKB1 Q921X6
A0A194PMD1 A0A194RLC3 I4DPJ5 A0A212ETQ9 A0A067R809 A0A2J7R275 A0A2J7R283 A0A0N0U431 A0A232ETH4 K7J1D6 A0A2A3EQF8 E9IUD9 A0A0L7R1J8 A0A087ZZS0 A0A026WU45 A0A154P4N9 A0A3L8D8Z5 A0A151WH64 E2AN67 A0A3L8D8H1 E2BG67 E0W056 A0A1L8DH24 A0A158NA11 A0A1Y1KIJ9 U5ELI2 A0A1I8P898 J3JTQ9 N6TQ48 A0A1B6LXI3 F4WC50 A0A1I8MSH0 A0A1B6DXE9 A0A195FQZ6 W8AF96 A0A1B6HCU6 A0A084W9B5 A0A182GJQ4 A0A023EP59 A0A182Q5K6 A0A0K8UN47 A0A151J0N8 A0A0L0BMZ5 D6WU15 A0A034WW18 S4RLM9 Q17EJ2 B0XD81 A0A1W4X6H6 A0A2R5L7G7 A0A182K081 C3XZ17 A0A1Q3F687 Q7PZL8 A0A195CUS1 A0A0L8FIU6 A0A182YBN2 A0A2K6EI85 A0A182L5B3 A0A182LSY9 G1T8N5 A0A182NZY6 A0A182W232 H0UYH0 K9IJ69 A0A2K6C5Y0 A0A2K6K0B3 A0A2K5XNF2 A0A0D9RGH0 A0A096NVN9 A0A2K6QUU0 A0A2K5KZY0 G7PH18 G7N320 A0A131XXX8 A0A1U7TY73 Q2T9S3 G3R7C7 G1RFC5 F6RMB3 A0A2R9BKK2 H2P144 H2QK06 Q9H1D9 H3ALW7 A0A147BQU5 D2HSX3 L8J1H4 W5NQ69 F1MYX7 E2RG78 A0A3Q7SCX0 A8QKB1 Q921X6
PDB
6F44
E-value=1.64589e-25,
Score=286
Ontologies
GO
PANTHER
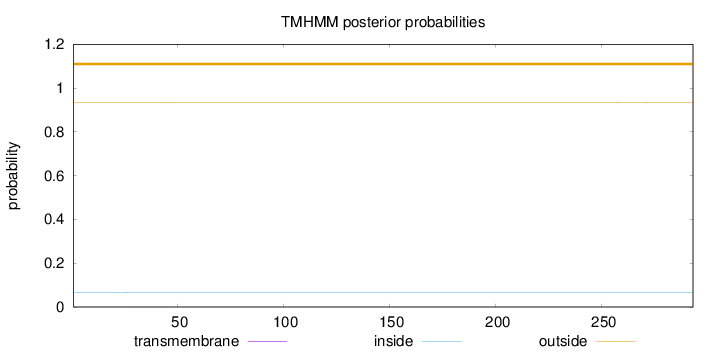
Topology
Subcellular location
Nucleus
Length:
293
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00903
Exp number, first 60 AAs:
0.00667
Total prob of N-in:
0.06587
outside
1 - 293
Population Genetic Test Statistics
Pi
265.768016
Theta
196.74905
Tajima's D
1.171557
CLR
0
CSRT
0.711814409279536
Interpretation
Uncertain
