Gene
KWMTBOMO15149
Pre Gene Modal
BGIBMGA004928
Annotation
PREDICTED:_uncharacterized_protein_C15orf41_homolog_[Bombyx_mori]
Full name
Uncharacterized protein C15orf41 homolog
Location in the cell
Cytoplasmic Reliability : 1.334 Mitochondrial Reliability : 1.141 Nuclear Reliability : 1.31
Sequence
CDS
ATGGACTTCCACGCAGGACAAACAAAACCGATGAAAATAGAAGAATACAATGCAATAGTGGAAGATTTTACTAAACTTTCATCATTCACTAGAAAAATGGAAAACGAACTTAAAAGAAAATATTGTCATATACAACCTCATACCTTGGGGAGTATCATATCATTATTAATACAAAGATCTATGAAACAGAACTATAGAAAATCCCCTGCAATATCATCTAAATATCAAGAGCTTTATGAAGTTGCTATGAGAGATGATACCAAAGAAAATGTTATCTTAAAGCTCTCTGAGAGTCAAGGTATCAGTCCAGCACTTTTTGCTAGGTCATTGCTACAAGGAGTGTTCTCAGACTCAACTATATCAAAGAAATATATTAAGGACACAACATTGATTGACAATAAAGATCTAGCTTACCAAGTTTTTATGGGAATTATGAATGACAATCAATATGGACCTTATGCGGATGTGACAAAGCAGTCAATTGGCTTGGAGTATGAGTTGAGACTAGAACGAGAGTTAAGGTTAATGAATATTACCTTTTCTGATGAAAATGTGCTACGTTCAAGAGGGTATGATAAAACTCCTGACTTCAAATTGGATGTACCCATCGCTGTTGATGGTTTTGTTATCAATTGGGTTGAAAGCAAAGCTTTATTTGGGGATGAAGAAAATCATTCTGGTTATCTTAAAGAACAGTTATTGTGTTACTGGAACAGATTTGGACCTGGTTTAGTGATTTATTGGTTTGGTTATTTAGAAACATTAGAATCTACTCCAGAAGTAAATAAAATGTTCATATTACGTACAAGTTTTCCAGATAAATCTAGCATAACTCAATATAAACCAGATATATTATAA
Protein
MDFHAGQTKPMKIEEYNAIVEDFTKLSSFTRKMENELKRKYCHIQPHTLGSIISLLIQRSMKQNYRKSPAISSKYQELYEVAMRDDTKENVILKLSESQGISPALFARSLLQGVFSDSTISKKYIKDTTLIDNKDLAYQVFMGIMNDNQYGPYADVTKQSIGLEYELRLERELRLMNITFSDENVLRSRGYDKTPDFKLDVPIAVDGFVINWVESKALFGDEENHSGYLKEQLLCYWNRFGPGLVIYWFGYLETLESTPEVNKMFILRTSFPDKSSITQYKPDIL
Summary
Keywords
Complete proteome
Reference proteome
Feature
chain Uncharacterized protein C15orf41 homolog
Uniprot
A0A2A4JML7
A0A2H1WZP2
A0A0L7K4I3
H9J5Y7
S4PA31
A0A194RLM8
+ More
A0A1E1WRW7 A0A212ETV2 A0A194PGV5 A0A067R5H2 A0A0L7QTS2 A0A2J7QRZ9 A0A088AB30 A0A2A3ERV8 A0A026WWP5 A0A1B6G5Q3 A0A195F7P4 A0A195AW95 A0A158NI07 F4WPT9 A0A195EH37 A0A195CLF4 A0A151WR21 E2AZW3 A0A0P4WGK2 A0A154P582 A0A1B6IAV3 A0A0T6BGZ3 E9IBJ8 A0A1B6ENG5 K7J7P1 A0A232F6D7 T1JCP0 D6WU13 E2BA56 E9HMN8 A0A1W4XGV3 A0A2J7QRZ4 A0A293M0D6 A0A0P4Y1M6 A0A0P6HHL9 A0A0P6E2Q2 A0A2J7QS10 A0A1B6FXR2 A0A2P8YG51 A0A1B6CVW3 A0A0P5X6V5 A0A224YMV5 A0A0P6F650 A0A131Z332 A7RP44 L7LZ70 A0A1S3J3H9 A0A1S3K9J4 A0A2J7QS07 A0A0J7KU12 A0A210QUT3 N6TV09 V5HRN3 A0A1E1X4X3 A0A1Y1MR97 B7P753 L7LS45 A0A2D0RRH7 A0A0L8HY00 G3MPE7 V4ACB0 W5L5P4 A0A3B1JEM9 W5N328 A0A1D2NNI4 A0A2D4G8D0 A0A146X7F1 A0A2B4STE6 S4RN60 A0A1W4Y4A4 A0A3Q2CBK7 A0A0F7ZEI8 A0A3B4BUU9 A0A3P9NMA1 A0A1A8CZ87 A0A1A8ANK1 Q5E9S8 G3P8I8 A0A1A8LBA3 A0A3M6T599 A0A1S3A1K9 A0A1S3KA30 M4A564 A0A3Q1I6E3 A0A1A8G1T6 A0A087YAR7 A0A1A8Q2A6 A0A3Q7RI81 A0A2Y9HHD3 A0A2U3WLE2 A0A1S3J3G3 W5QFJ3 A0A1A8HUG1
A0A1E1WRW7 A0A212ETV2 A0A194PGV5 A0A067R5H2 A0A0L7QTS2 A0A2J7QRZ9 A0A088AB30 A0A2A3ERV8 A0A026WWP5 A0A1B6G5Q3 A0A195F7P4 A0A195AW95 A0A158NI07 F4WPT9 A0A195EH37 A0A195CLF4 A0A151WR21 E2AZW3 A0A0P4WGK2 A0A154P582 A0A1B6IAV3 A0A0T6BGZ3 E9IBJ8 A0A1B6ENG5 K7J7P1 A0A232F6D7 T1JCP0 D6WU13 E2BA56 E9HMN8 A0A1W4XGV3 A0A2J7QRZ4 A0A293M0D6 A0A0P4Y1M6 A0A0P6HHL9 A0A0P6E2Q2 A0A2J7QS10 A0A1B6FXR2 A0A2P8YG51 A0A1B6CVW3 A0A0P5X6V5 A0A224YMV5 A0A0P6F650 A0A131Z332 A7RP44 L7LZ70 A0A1S3J3H9 A0A1S3K9J4 A0A2J7QS07 A0A0J7KU12 A0A210QUT3 N6TV09 V5HRN3 A0A1E1X4X3 A0A1Y1MR97 B7P753 L7LS45 A0A2D0RRH7 A0A0L8HY00 G3MPE7 V4ACB0 W5L5P4 A0A3B1JEM9 W5N328 A0A1D2NNI4 A0A2D4G8D0 A0A146X7F1 A0A2B4STE6 S4RN60 A0A1W4Y4A4 A0A3Q2CBK7 A0A0F7ZEI8 A0A3B4BUU9 A0A3P9NMA1 A0A1A8CZ87 A0A1A8ANK1 Q5E9S8 G3P8I8 A0A1A8LBA3 A0A3M6T599 A0A1S3A1K9 A0A1S3KA30 M4A564 A0A3Q1I6E3 A0A1A8G1T6 A0A087YAR7 A0A1A8Q2A6 A0A3Q7RI81 A0A2Y9HHD3 A0A2U3WLE2 A0A1S3J3G3 W5QFJ3 A0A1A8HUG1
Pubmed
26227816
19121390
23622113
26354079
22118469
24845553
+ More
24508170 30249741 21347285 21719571 20798317 21282665 20075255 28648823 18362917 19820115 21292972 29403074 28797301 26830274 17615350 25576852 26383154 28812685 23537049 25765539 28503490 28004739 22216098 23254933 25329095 27289101 16305752 30382153 23542700 20809919
24508170 30249741 21347285 21719571 20798317 21282665 20075255 28648823 18362917 19820115 21292972 29403074 28797301 26830274 17615350 25576852 26383154 28812685 23537049 25765539 28503490 28004739 22216098 23254933 25329095 27289101 16305752 30382153 23542700 20809919
EMBL
NWSH01001030
PCG73026.1
ODYU01012229
SOQ58426.1
JTDY01011012
KOB57703.1
+ More
BABH01029966 BABH01029967 GAIX01003434 JAA89126.1 KQ460045 KPJ18220.1 GDQN01001332 JAT89722.1 AGBW02012531 OWR44881.1 KQ459604 KPI92273.1 KK852730 KDR17515.1 KQ414741 KOC61954.1 NEVH01011882 PNF31359.1 KZ288194 PBC33859.1 KK107078 QOIP01000011 EZA60248.1 RLU16893.1 GECZ01011999 JAS57770.1 KQ981763 KYN36084.1 KQ976725 KYM76518.1 ADTU01016132 GL888255 EGI63768.1 KQ978957 KYN27179.1 KQ977622 KYN01297.1 KQ982813 KYQ50349.1 GL444280 EFN61012.1 GDRN01090228 JAI60502.1 KQ434809 KZC06338.1 GECU01023683 JAS84023.1 LJIG01000327 KRT86613.1 GL762111 EFZ22162.1 GECZ01030323 JAS39446.1 AAZX01015390 NNAY01000808 OXU26404.1 JH432074 KQ971352 EFA07300.1 GL446664 EFN87423.1 GL732688 EFX67010.1 PNF31358.1 GFWV01007735 MAA32465.1 GDIP01243260 JAI80141.1 GDIQ01028017 JAN66720.1 GDIQ01068624 JAN26113.1 PNF31356.1 GECZ01014784 JAS54985.1 PYGN01000627 PSN43164.1 GEDC01019955 JAS17343.1 GDIP01076564 JAM27151.1 GFPF01004755 MAA15901.1 GDIQ01052882 JAN41855.1 GEDV01003365 JAP85192.1 DS469524 EDO46791.1 GACK01007969 JAA57065.1 PNF31355.1 LBMM01003259 KMQ93739.1 NEDP02001786 OWF52494.1 APGK01051081 KB741181 ENN73085.1 GANP01003594 JAB80874.1 GFAC01004884 JAT94304.1 GEZM01023877 GEZM01023875 JAV88181.1 ABJB010540763 DS649438 EEC02425.1 GACK01010494 JAA54540.1 KQ417037 KOF94071.1 JO843748 AEO35365.1 KB202014 ESO92740.1 AHAT01019067 LJIJ01000001 ODN06645.1 IACJ01110759 LAA55997.1 GCES01048272 GCES01010254 JAR38051.1 LSMT01000024 PFX32379.1 GBEX01000441 JAI14119.1 HADZ01020402 SBP84343.1 HADY01017591 HAEJ01008440 SBP56076.1 BT020842 BC142022 HAEF01003782 SBR41164.1 RCHS01004322 RMX36459.1 HAEB01018474 SBQ65001.1 AYCK01001357 HAEH01009621 SBR87626.1 AMGL01102537 AMGL01102538 AMGL01102539 AMGL01102540 AMGL01102541 AMGL01102542 AMGL01102543 HAED01003019 SBQ88896.1
BABH01029966 BABH01029967 GAIX01003434 JAA89126.1 KQ460045 KPJ18220.1 GDQN01001332 JAT89722.1 AGBW02012531 OWR44881.1 KQ459604 KPI92273.1 KK852730 KDR17515.1 KQ414741 KOC61954.1 NEVH01011882 PNF31359.1 KZ288194 PBC33859.1 KK107078 QOIP01000011 EZA60248.1 RLU16893.1 GECZ01011999 JAS57770.1 KQ981763 KYN36084.1 KQ976725 KYM76518.1 ADTU01016132 GL888255 EGI63768.1 KQ978957 KYN27179.1 KQ977622 KYN01297.1 KQ982813 KYQ50349.1 GL444280 EFN61012.1 GDRN01090228 JAI60502.1 KQ434809 KZC06338.1 GECU01023683 JAS84023.1 LJIG01000327 KRT86613.1 GL762111 EFZ22162.1 GECZ01030323 JAS39446.1 AAZX01015390 NNAY01000808 OXU26404.1 JH432074 KQ971352 EFA07300.1 GL446664 EFN87423.1 GL732688 EFX67010.1 PNF31358.1 GFWV01007735 MAA32465.1 GDIP01243260 JAI80141.1 GDIQ01028017 JAN66720.1 GDIQ01068624 JAN26113.1 PNF31356.1 GECZ01014784 JAS54985.1 PYGN01000627 PSN43164.1 GEDC01019955 JAS17343.1 GDIP01076564 JAM27151.1 GFPF01004755 MAA15901.1 GDIQ01052882 JAN41855.1 GEDV01003365 JAP85192.1 DS469524 EDO46791.1 GACK01007969 JAA57065.1 PNF31355.1 LBMM01003259 KMQ93739.1 NEDP02001786 OWF52494.1 APGK01051081 KB741181 ENN73085.1 GANP01003594 JAB80874.1 GFAC01004884 JAT94304.1 GEZM01023877 GEZM01023875 JAV88181.1 ABJB010540763 DS649438 EEC02425.1 GACK01010494 JAA54540.1 KQ417037 KOF94071.1 JO843748 AEO35365.1 KB202014 ESO92740.1 AHAT01019067 LJIJ01000001 ODN06645.1 IACJ01110759 LAA55997.1 GCES01048272 GCES01010254 JAR38051.1 LSMT01000024 PFX32379.1 GBEX01000441 JAI14119.1 HADZ01020402 SBP84343.1 HADY01017591 HAEJ01008440 SBP56076.1 BT020842 BC142022 HAEF01003782 SBR41164.1 RCHS01004322 RMX36459.1 HAEB01018474 SBQ65001.1 AYCK01001357 HAEH01009621 SBR87626.1 AMGL01102537 AMGL01102538 AMGL01102539 AMGL01102540 AMGL01102541 AMGL01102542 AMGL01102543 HAED01003019 SBQ88896.1
Proteomes
UP000218220
UP000037510
UP000005204
UP000053240
UP000007151
UP000053268
+ More
UP000027135 UP000053825 UP000235965 UP000005203 UP000242457 UP000053097 UP000279307 UP000078541 UP000078540 UP000005205 UP000007755 UP000078492 UP000078542 UP000075809 UP000000311 UP000076502 UP000002358 UP000215335 UP000007266 UP000008237 UP000000305 UP000192223 UP000245037 UP000001593 UP000085678 UP000036403 UP000242188 UP000019118 UP000001555 UP000221080 UP000053454 UP000030746 UP000018467 UP000018468 UP000094527 UP000265000 UP000225706 UP000245300 UP000192224 UP000265020 UP000261440 UP000242638 UP000009136 UP000007635 UP000275408 UP000079721 UP000002852 UP000265040 UP000028760 UP000286641 UP000248481 UP000245340 UP000002356
UP000027135 UP000053825 UP000235965 UP000005203 UP000242457 UP000053097 UP000279307 UP000078541 UP000078540 UP000005205 UP000007755 UP000078492 UP000078542 UP000075809 UP000000311 UP000076502 UP000002358 UP000215335 UP000007266 UP000008237 UP000000305 UP000192223 UP000245037 UP000001593 UP000085678 UP000036403 UP000242188 UP000019118 UP000001555 UP000221080 UP000053454 UP000030746 UP000018467 UP000018468 UP000094527 UP000265000 UP000225706 UP000245300 UP000192224 UP000265020 UP000261440 UP000242638 UP000009136 UP000007635 UP000275408 UP000079721 UP000002852 UP000265040 UP000028760 UP000286641 UP000248481 UP000245340 UP000002356
Pfam
PF14811 TPD
Interpro
IPR029404
DUF_TPD
ProteinModelPortal
A0A2A4JML7
A0A2H1WZP2
A0A0L7K4I3
H9J5Y7
S4PA31
A0A194RLM8
+ More
A0A1E1WRW7 A0A212ETV2 A0A194PGV5 A0A067R5H2 A0A0L7QTS2 A0A2J7QRZ9 A0A088AB30 A0A2A3ERV8 A0A026WWP5 A0A1B6G5Q3 A0A195F7P4 A0A195AW95 A0A158NI07 F4WPT9 A0A195EH37 A0A195CLF4 A0A151WR21 E2AZW3 A0A0P4WGK2 A0A154P582 A0A1B6IAV3 A0A0T6BGZ3 E9IBJ8 A0A1B6ENG5 K7J7P1 A0A232F6D7 T1JCP0 D6WU13 E2BA56 E9HMN8 A0A1W4XGV3 A0A2J7QRZ4 A0A293M0D6 A0A0P4Y1M6 A0A0P6HHL9 A0A0P6E2Q2 A0A2J7QS10 A0A1B6FXR2 A0A2P8YG51 A0A1B6CVW3 A0A0P5X6V5 A0A224YMV5 A0A0P6F650 A0A131Z332 A7RP44 L7LZ70 A0A1S3J3H9 A0A1S3K9J4 A0A2J7QS07 A0A0J7KU12 A0A210QUT3 N6TV09 V5HRN3 A0A1E1X4X3 A0A1Y1MR97 B7P753 L7LS45 A0A2D0RRH7 A0A0L8HY00 G3MPE7 V4ACB0 W5L5P4 A0A3B1JEM9 W5N328 A0A1D2NNI4 A0A2D4G8D0 A0A146X7F1 A0A2B4STE6 S4RN60 A0A1W4Y4A4 A0A3Q2CBK7 A0A0F7ZEI8 A0A3B4BUU9 A0A3P9NMA1 A0A1A8CZ87 A0A1A8ANK1 Q5E9S8 G3P8I8 A0A1A8LBA3 A0A3M6T599 A0A1S3A1K9 A0A1S3KA30 M4A564 A0A3Q1I6E3 A0A1A8G1T6 A0A087YAR7 A0A1A8Q2A6 A0A3Q7RI81 A0A2Y9HHD3 A0A2U3WLE2 A0A1S3J3G3 W5QFJ3 A0A1A8HUG1
A0A1E1WRW7 A0A212ETV2 A0A194PGV5 A0A067R5H2 A0A0L7QTS2 A0A2J7QRZ9 A0A088AB30 A0A2A3ERV8 A0A026WWP5 A0A1B6G5Q3 A0A195F7P4 A0A195AW95 A0A158NI07 F4WPT9 A0A195EH37 A0A195CLF4 A0A151WR21 E2AZW3 A0A0P4WGK2 A0A154P582 A0A1B6IAV3 A0A0T6BGZ3 E9IBJ8 A0A1B6ENG5 K7J7P1 A0A232F6D7 T1JCP0 D6WU13 E2BA56 E9HMN8 A0A1W4XGV3 A0A2J7QRZ4 A0A293M0D6 A0A0P4Y1M6 A0A0P6HHL9 A0A0P6E2Q2 A0A2J7QS10 A0A1B6FXR2 A0A2P8YG51 A0A1B6CVW3 A0A0P5X6V5 A0A224YMV5 A0A0P6F650 A0A131Z332 A7RP44 L7LZ70 A0A1S3J3H9 A0A1S3K9J4 A0A2J7QS07 A0A0J7KU12 A0A210QUT3 N6TV09 V5HRN3 A0A1E1X4X3 A0A1Y1MR97 B7P753 L7LS45 A0A2D0RRH7 A0A0L8HY00 G3MPE7 V4ACB0 W5L5P4 A0A3B1JEM9 W5N328 A0A1D2NNI4 A0A2D4G8D0 A0A146X7F1 A0A2B4STE6 S4RN60 A0A1W4Y4A4 A0A3Q2CBK7 A0A0F7ZEI8 A0A3B4BUU9 A0A3P9NMA1 A0A1A8CZ87 A0A1A8ANK1 Q5E9S8 G3P8I8 A0A1A8LBA3 A0A3M6T599 A0A1S3A1K9 A0A1S3KA30 M4A564 A0A3Q1I6E3 A0A1A8G1T6 A0A087YAR7 A0A1A8Q2A6 A0A3Q7RI81 A0A2Y9HHD3 A0A2U3WLE2 A0A1S3J3G3 W5QFJ3 A0A1A8HUG1
Ontologies
GO
PANTHER
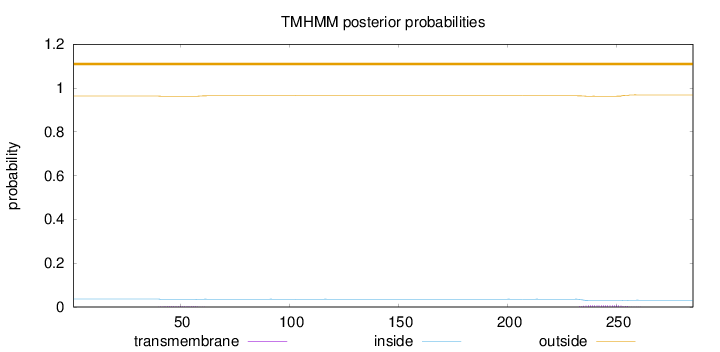
Topology
Length:
285
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.20149
Exp number, first 60 AAs:
0.04948
Total prob of N-in:
0.03681
outside
1 - 285
Population Genetic Test Statistics
Pi
254.447163
Theta
197.01938
Tajima's D
0.917436
CLR
0.000674
CSRT
0.63816809159542
Interpretation
Uncertain
