Gene
KWMTBOMO15145
Pre Gene Modal
BGIBMGA004986
Annotation
PREDICTED:_protein_farnesyltransferase_subunit_beta_[Bombyx_mori]
Full name
Protein farnesyltransferase subunit beta
Location in the cell
Cytoplasmic Reliability : 1.259 Extracellular Reliability : 1.581
Sequence
CDS
ATGGAAAACCAAGTCCGTTGTTACAACGATATTGTATCGGAAGCTTTCGATTCTGAAGATGTTAAAACAGACACATCAATAGATCAGATTTATGTTGAAAAAACAATTTTGAAATTGTACAAGGAATTTGAGAAAAATTCCTCCATTGATCCTGATTTACCACTTTTGATAAAAGAAGTTCACAGCAAGCTATTAAAAAAATGTTTAATCAATTTGCCAAAAAGTTATGAATGCCTTGATGCTAGCAGAACCTGGATCATATATTGGATTCTACATTCACTATGGATTCTTAATGACATGCCCGACCATGAGACTCTTTCTGCAGTAGTAAAATTCTTAGATCAGTGTCAACATGAAGATGGCGGTTATGGTGGGGGGCCTAGACAATATCCACATTTGGGAACAACATATGCAGCTGTTAACGCCCTTAGCATAATAGGAACAGATGAGGCTTATGACTCAATAGATAGAAGTAGCTTACAAAGATTTCTGTGGACAGTTAGAGATGTTGATGGATCCTTTGCTCTTCATAAAGATGGAGAACAAGATATTAGAGGCGCATATTGTGCAATTAGTATAGCCAAAATGACAAATACATACACAGAAGCGTTATTTGATAAAACGGCAGAATGGATTGTTAGTTGTCAGACATATGAGGGTGGCTTTGCTGGTTGTCCTGGTATGGAAGCACATGGTGGCTATGCATTCTGTGGAATCGCATCCTTAGCTCTATTAAATAGAACACAATTATGTGATATAGATTCACTGCTTAGGTGGTCAGTAAACAGACAGATGAGGATAGAGGGTGGATTTCAAGGAAGGACAAACAAACTTGTAGATGGATGTTATTCTTTTTGGCAAGGTGCTGCTTTTCCTATTATAAGTGCAATACTTTCAAAAGATAATAAGGAACTTATTGAGACAGTTCTTTTCAATCAATCTGCCCTACAGGAATACATTTTGATATGTTGTCAAAATCGGGATGGTGGTCTGATTGATAAGCCTGGCAAGCCTAGAGACATTTACCATACATGTTATGGCCTCAGTGGATTATCGGTAGCGCAACATGGTACAGGGGTTATGGATCCCTACGTAGTTGGATCACCTAGTAATGAACTGAATAGAGTGCATCCTTTGCATAATGTTGCACCTCATTTGGTTTACAACAGTGTTCACTATTTTATCAGGCACCCGCCTCCAGTCAAAGACAAGAATTAA
Protein
MENQVRCYNDIVSEAFDSEDVKTDTSIDQIYVEKTILKLYKEFEKNSSIDPDLPLLIKEVHSKLLKKCLINLPKSYECLDASRTWIIYWILHSLWILNDMPDHETLSAVVKFLDQCQHEDGGYGGGPRQYPHLGTTYAAVNALSIIGTDEAYDSIDRSSLQRFLWTVRDVDGSFALHKDGEQDIRGAYCAISIAKMTNTYTEALFDKTAEWIVSCQTYEGGFAGCPGMEAHGGYAFCGIASLALLNRTQLCDIDSLLRWSVNRQMRIEGGFQGRTNKLVDGCYSFWQGAAFPIISAILSKDNKELIETVLFNQSALQEYILICCQNRDGGLIDKPGKPRDIYHTCYGLSGLSVAQHGTGVMDPYVVGSPSNELNRVHPLHNVAPHLVYNSVHYFIRHPPPVKDKN
Summary
Description
Catalyzes the transfer of a farnesyl moiety from farnesyl diphosphate to a cysteine at the fourth position from the C-terminus of several proteins. The beta subunit is responsible for peptide-binding.
Catalytic Activity
(2E,6E)-farnesyl diphosphate + L-cysteinyl-[protein] = diphosphate + S-(2E,6E)-farnesyl-L-cysteinyl-[protein]
Cofactor
Zn(2+)
Subunit
Heterodimer of an alpha and a beta subunit.
Similarity
Belongs to the protein prenyltransferase subunit beta family.
Belongs to the cyclin family.
Belongs to the cyclin family.
Feature
chain Protein farnesyltransferase subunit beta
Uniprot
E2QC37
A0A2A4JM34
A0A2H1WZI3
A0A212ETQ7
A0A1E1WE49
A0A194PGV1
+ More
A0A0L7KSY6 A0A194RLC8 A0A067R136 A0A2J7QAW9 A0A1W4XHW9 A0A0L0CJJ2 A0A1B6CPL5 A0A336MDZ1 A0A1Y1L156 A0A1A9WH29 D3TP88 A0A1A9YCT7 A0A1A9Z3U9 A0A1B0G9M1 A0A1B6HD43 A0A1B0BUH6 A0A0A1WJA9 A0A034V7Z1 A0A0K8UKP2 A0A1B6EPX8 A0A293N1C3 A0A224Z2A8 R7UM01 T1PC76 A0A1B6L534 W8BQ94 A0A131YYF3 A0A2R5LKX7 A0A1B6DT40 L7M4M2 A0A1S3HPM9 A0A2R2WSJ4 A0A1I8P731 A0A1Z5LCF7 D6WSN3 A0A1A9V7X0 E2ANE8 A0A151IQ29 A0A2H8TSE4 E0VV79 A0A2G8LR97 A0A2S2N6U6 J3JUW9 V3YZC1 E9IZL7 A0A151I3F7 A0A1J1IWT7 A0A151J5R8 A0A158NIN8 J9K4H2 B4LWX0 U4U0Q3 N6TVJ2 B4KBZ8 A0A2S2R6M8 A0A232F293 A0A131XVC9 A0A147BQI4 A0A195FS92 A0A1Q3G1T2 A0A3L8DJT5 A0A026WM91 A0A2C9L7R8 A0A182QI73 A0A1E1X5D9 I3JEI6 A0A182PSH7 A0A0M4F6L4 A0A1L8DTT6 E2BEM9 A0A3B4EQF7 A0A3Q2VTU3 A0A3P8RBP7 A0A3P9B2C7 A0A084VXQ9 A0A3Q4FZZ4 U5EHI4 A0A0P4W3G2 A0A088AFG0 Q4RQW7 A0A0P4Z5Y7 A0A0P6EEN8 A0A0P5T528 A0A0P5RLT1 A0A182NRS6 H3DG66 A0A310SCJ8 A0A164T713 A0A2M4APS2 A0A3S1B2Q5 B4IBW2 A0A182VBP7 B4QX36 A0A2M4AQ49
A0A0L7KSY6 A0A194RLC8 A0A067R136 A0A2J7QAW9 A0A1W4XHW9 A0A0L0CJJ2 A0A1B6CPL5 A0A336MDZ1 A0A1Y1L156 A0A1A9WH29 D3TP88 A0A1A9YCT7 A0A1A9Z3U9 A0A1B0G9M1 A0A1B6HD43 A0A1B0BUH6 A0A0A1WJA9 A0A034V7Z1 A0A0K8UKP2 A0A1B6EPX8 A0A293N1C3 A0A224Z2A8 R7UM01 T1PC76 A0A1B6L534 W8BQ94 A0A131YYF3 A0A2R5LKX7 A0A1B6DT40 L7M4M2 A0A1S3HPM9 A0A2R2WSJ4 A0A1I8P731 A0A1Z5LCF7 D6WSN3 A0A1A9V7X0 E2ANE8 A0A151IQ29 A0A2H8TSE4 E0VV79 A0A2G8LR97 A0A2S2N6U6 J3JUW9 V3YZC1 E9IZL7 A0A151I3F7 A0A1J1IWT7 A0A151J5R8 A0A158NIN8 J9K4H2 B4LWX0 U4U0Q3 N6TVJ2 B4KBZ8 A0A2S2R6M8 A0A232F293 A0A131XVC9 A0A147BQI4 A0A195FS92 A0A1Q3G1T2 A0A3L8DJT5 A0A026WM91 A0A2C9L7R8 A0A182QI73 A0A1E1X5D9 I3JEI6 A0A182PSH7 A0A0M4F6L4 A0A1L8DTT6 E2BEM9 A0A3B4EQF7 A0A3Q2VTU3 A0A3P8RBP7 A0A3P9B2C7 A0A084VXQ9 A0A3Q4FZZ4 U5EHI4 A0A0P4W3G2 A0A088AFG0 Q4RQW7 A0A0P4Z5Y7 A0A0P6EEN8 A0A0P5T528 A0A0P5RLT1 A0A182NRS6 H3DG66 A0A310SCJ8 A0A164T713 A0A2M4APS2 A0A3S1B2Q5 B4IBW2 A0A182VBP7 B4QX36 A0A2M4AQ49
EC Number
2.5.1.58
Pubmed
19121390
22118469
26354079
26227816
24845553
26108605
+ More
28004739 20353571 25830018 25348373 28797301 23254933 25315136 24495485 26830274 25576852 28528879 18362917 19820115 20798317 20566863 29023486 22516182 21282665 21347285 17994087 23537049 28648823 29652888 30249741 24508170 15562597 28503490 25186727 24438588 15496914
28004739 20353571 25830018 25348373 28797301 23254933 25315136 24495485 26830274 25576852 28528879 18362917 19820115 20798317 20566863 29023486 22516182 21282665 21347285 17994087 23537049 28648823 29652888 30249741 24508170 15562597 28503490 25186727 24438588 15496914
EMBL
BABH01029968
BABH01029969
AB201556
BAD89510.1
NWSH01001030
PCG73041.1
+ More
ODYU01012229 SOQ58428.1 AGBW02012531 OWR44885.1 GDQN01005923 JAT85131.1 KQ459604 KPI92268.1 JTDY01006181 KOB66215.1 KQ460045 KPJ18224.1 KK853013 KDR12520.1 NEVH01016310 PNF25731.1 JRES01000301 KNC32583.1 GEDC01021928 JAS15370.1 UFQT01000693 SSX26607.1 GEZM01067832 JAV67393.1 EZ423240 ADD19516.1 CCAG010022856 GECU01035103 JAS72603.1 JXJN01020692 GBXI01015148 JAC99143.1 GAKP01019526 JAC39426.1 GDHF01025178 JAI27136.1 GECZ01029772 JAS39997.1 GFWV01021441 MAA46169.1 GFPF01010953 MAA22099.1 AMQN01007169 KB300141 ELU07098.1 KA645538 AFP60167.1 GEBQ01021236 JAT18741.1 GAMC01007582 GAMC01007580 JAB98973.1 GEDV01004338 JAP84219.1 GGLE01006054 MBY10180.1 GEDC01008473 JAS28825.1 GACK01006961 JAA58073.1 KY628458 ARE68914.1 GFJQ02001975 JAW04995.1 KQ971354 EFA07131.1 GL441193 EFN65048.1 KQ976794 KYN08259.1 GFXV01004707 MBW16512.1 DS235804 EEB17285.1 MRZV01000007 PIK62700.1 GGMR01000179 MBY12798.1 BT127034 AEE61996.1 KB203660 ESO83518.1 GL767232 EFZ13918.1 KQ976479 KYM83776.1 CVRI01000059 CRL03017.1 KQ979945 KYN18574.1 ADTU01017033 ABLF02028179 CH940650 EDW66691.1 KB631951 ERL87434.1 APGK01016859 APGK01016860 APGK01016861 APGK01016862 APGK01016863 KB739988 ENN82053.1 CH933806 EDW15850.1 GGMS01016452 MBY85655.1 NNAY01001181 OXU24831.1 GEFM01004543 JAP71253.1 GEGO01002632 JAR92772.1 KQ981285 KYN43167.1 GFDL01001292 JAV33753.1 QOIP01000007 RLU20657.1 KK107152 EZA57177.1 AXCN02001149 GFAC01004734 JAT94454.1 AERX01015206 CP012526 ALC47712.1 GFDF01004205 JAV09879.1 GL447829 EFN85863.1 ATLV01018182 KE525221 KFB42753.1 GANO01003009 JAB56862.1 GDRN01092204 JAI60150.1 CAAE01015003 CAG09215.1 GDIP01219978 JAJ03424.1 GDIQ01063705 JAN31032.1 GDIQ01096446 JAL55280.1 GDIQ01098945 GDIP01086614 JAL52781.1 JAM17101.1 KQ777939 OAD52037.1 LRGB01001867 KZS10239.1 GGFK01009463 MBW42784.1 RQTK01000919 RUS73593.1 CH480827 EDW44870.1 CM000364 EDX12690.1 GGFK01009417 MBW42738.1
ODYU01012229 SOQ58428.1 AGBW02012531 OWR44885.1 GDQN01005923 JAT85131.1 KQ459604 KPI92268.1 JTDY01006181 KOB66215.1 KQ460045 KPJ18224.1 KK853013 KDR12520.1 NEVH01016310 PNF25731.1 JRES01000301 KNC32583.1 GEDC01021928 JAS15370.1 UFQT01000693 SSX26607.1 GEZM01067832 JAV67393.1 EZ423240 ADD19516.1 CCAG010022856 GECU01035103 JAS72603.1 JXJN01020692 GBXI01015148 JAC99143.1 GAKP01019526 JAC39426.1 GDHF01025178 JAI27136.1 GECZ01029772 JAS39997.1 GFWV01021441 MAA46169.1 GFPF01010953 MAA22099.1 AMQN01007169 KB300141 ELU07098.1 KA645538 AFP60167.1 GEBQ01021236 JAT18741.1 GAMC01007582 GAMC01007580 JAB98973.1 GEDV01004338 JAP84219.1 GGLE01006054 MBY10180.1 GEDC01008473 JAS28825.1 GACK01006961 JAA58073.1 KY628458 ARE68914.1 GFJQ02001975 JAW04995.1 KQ971354 EFA07131.1 GL441193 EFN65048.1 KQ976794 KYN08259.1 GFXV01004707 MBW16512.1 DS235804 EEB17285.1 MRZV01000007 PIK62700.1 GGMR01000179 MBY12798.1 BT127034 AEE61996.1 KB203660 ESO83518.1 GL767232 EFZ13918.1 KQ976479 KYM83776.1 CVRI01000059 CRL03017.1 KQ979945 KYN18574.1 ADTU01017033 ABLF02028179 CH940650 EDW66691.1 KB631951 ERL87434.1 APGK01016859 APGK01016860 APGK01016861 APGK01016862 APGK01016863 KB739988 ENN82053.1 CH933806 EDW15850.1 GGMS01016452 MBY85655.1 NNAY01001181 OXU24831.1 GEFM01004543 JAP71253.1 GEGO01002632 JAR92772.1 KQ981285 KYN43167.1 GFDL01001292 JAV33753.1 QOIP01000007 RLU20657.1 KK107152 EZA57177.1 AXCN02001149 GFAC01004734 JAT94454.1 AERX01015206 CP012526 ALC47712.1 GFDF01004205 JAV09879.1 GL447829 EFN85863.1 ATLV01018182 KE525221 KFB42753.1 GANO01003009 JAB56862.1 GDRN01092204 JAI60150.1 CAAE01015003 CAG09215.1 GDIP01219978 JAJ03424.1 GDIQ01063705 JAN31032.1 GDIQ01096446 JAL55280.1 GDIQ01098945 GDIP01086614 JAL52781.1 JAM17101.1 KQ777939 OAD52037.1 LRGB01001867 KZS10239.1 GGFK01009463 MBW42784.1 RQTK01000919 RUS73593.1 CH480827 EDW44870.1 CM000364 EDX12690.1 GGFK01009417 MBW42738.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000037510
UP000053240
+ More
UP000027135 UP000235965 UP000192223 UP000037069 UP000091820 UP000092443 UP000092445 UP000092444 UP000092460 UP000014760 UP000095301 UP000085678 UP000095300 UP000007266 UP000078200 UP000000311 UP000078542 UP000009046 UP000230750 UP000030746 UP000078540 UP000183832 UP000078492 UP000005205 UP000007819 UP000008792 UP000030742 UP000019118 UP000009192 UP000215335 UP000078541 UP000279307 UP000053097 UP000076420 UP000075886 UP000005207 UP000075885 UP000092553 UP000008237 UP000261460 UP000264840 UP000265100 UP000265160 UP000030765 UP000261580 UP000005203 UP000075884 UP000007303 UP000076858 UP000271974 UP000001292 UP000075903 UP000000304
UP000027135 UP000235965 UP000192223 UP000037069 UP000091820 UP000092443 UP000092445 UP000092444 UP000092460 UP000014760 UP000095301 UP000085678 UP000095300 UP000007266 UP000078200 UP000000311 UP000078542 UP000009046 UP000230750 UP000030746 UP000078540 UP000183832 UP000078492 UP000005205 UP000007819 UP000008792 UP000030742 UP000019118 UP000009192 UP000215335 UP000078541 UP000279307 UP000053097 UP000076420 UP000075886 UP000005207 UP000075885 UP000092553 UP000008237 UP000261460 UP000264840 UP000265100 UP000265160 UP000030765 UP000261580 UP000005203 UP000075884 UP000007303 UP000076858 UP000271974 UP000001292 UP000075903 UP000000304
PRIDE
Interpro
ProteinModelPortal
E2QC37
A0A2A4JM34
A0A2H1WZI3
A0A212ETQ7
A0A1E1WE49
A0A194PGV1
+ More
A0A0L7KSY6 A0A194RLC8 A0A067R136 A0A2J7QAW9 A0A1W4XHW9 A0A0L0CJJ2 A0A1B6CPL5 A0A336MDZ1 A0A1Y1L156 A0A1A9WH29 D3TP88 A0A1A9YCT7 A0A1A9Z3U9 A0A1B0G9M1 A0A1B6HD43 A0A1B0BUH6 A0A0A1WJA9 A0A034V7Z1 A0A0K8UKP2 A0A1B6EPX8 A0A293N1C3 A0A224Z2A8 R7UM01 T1PC76 A0A1B6L534 W8BQ94 A0A131YYF3 A0A2R5LKX7 A0A1B6DT40 L7M4M2 A0A1S3HPM9 A0A2R2WSJ4 A0A1I8P731 A0A1Z5LCF7 D6WSN3 A0A1A9V7X0 E2ANE8 A0A151IQ29 A0A2H8TSE4 E0VV79 A0A2G8LR97 A0A2S2N6U6 J3JUW9 V3YZC1 E9IZL7 A0A151I3F7 A0A1J1IWT7 A0A151J5R8 A0A158NIN8 J9K4H2 B4LWX0 U4U0Q3 N6TVJ2 B4KBZ8 A0A2S2R6M8 A0A232F293 A0A131XVC9 A0A147BQI4 A0A195FS92 A0A1Q3G1T2 A0A3L8DJT5 A0A026WM91 A0A2C9L7R8 A0A182QI73 A0A1E1X5D9 I3JEI6 A0A182PSH7 A0A0M4F6L4 A0A1L8DTT6 E2BEM9 A0A3B4EQF7 A0A3Q2VTU3 A0A3P8RBP7 A0A3P9B2C7 A0A084VXQ9 A0A3Q4FZZ4 U5EHI4 A0A0P4W3G2 A0A088AFG0 Q4RQW7 A0A0P4Z5Y7 A0A0P6EEN8 A0A0P5T528 A0A0P5RLT1 A0A182NRS6 H3DG66 A0A310SCJ8 A0A164T713 A0A2M4APS2 A0A3S1B2Q5 B4IBW2 A0A182VBP7 B4QX36 A0A2M4AQ49
A0A0L7KSY6 A0A194RLC8 A0A067R136 A0A2J7QAW9 A0A1W4XHW9 A0A0L0CJJ2 A0A1B6CPL5 A0A336MDZ1 A0A1Y1L156 A0A1A9WH29 D3TP88 A0A1A9YCT7 A0A1A9Z3U9 A0A1B0G9M1 A0A1B6HD43 A0A1B0BUH6 A0A0A1WJA9 A0A034V7Z1 A0A0K8UKP2 A0A1B6EPX8 A0A293N1C3 A0A224Z2A8 R7UM01 T1PC76 A0A1B6L534 W8BQ94 A0A131YYF3 A0A2R5LKX7 A0A1B6DT40 L7M4M2 A0A1S3HPM9 A0A2R2WSJ4 A0A1I8P731 A0A1Z5LCF7 D6WSN3 A0A1A9V7X0 E2ANE8 A0A151IQ29 A0A2H8TSE4 E0VV79 A0A2G8LR97 A0A2S2N6U6 J3JUW9 V3YZC1 E9IZL7 A0A151I3F7 A0A1J1IWT7 A0A151J5R8 A0A158NIN8 J9K4H2 B4LWX0 U4U0Q3 N6TVJ2 B4KBZ8 A0A2S2R6M8 A0A232F293 A0A131XVC9 A0A147BQI4 A0A195FS92 A0A1Q3G1T2 A0A3L8DJT5 A0A026WM91 A0A2C9L7R8 A0A182QI73 A0A1E1X5D9 I3JEI6 A0A182PSH7 A0A0M4F6L4 A0A1L8DTT6 E2BEM9 A0A3B4EQF7 A0A3Q2VTU3 A0A3P8RBP7 A0A3P9B2C7 A0A084VXQ9 A0A3Q4FZZ4 U5EHI4 A0A0P4W3G2 A0A088AFG0 Q4RQW7 A0A0P4Z5Y7 A0A0P6EEN8 A0A0P5T528 A0A0P5RLT1 A0A182NRS6 H3DG66 A0A310SCJ8 A0A164T713 A0A2M4APS2 A0A3S1B2Q5 B4IBW2 A0A182VBP7 B4QX36 A0A2M4AQ49
PDB
3E37
E-value=1.43518e-109,
Score=1013
Ontologies
GO
PANTHER
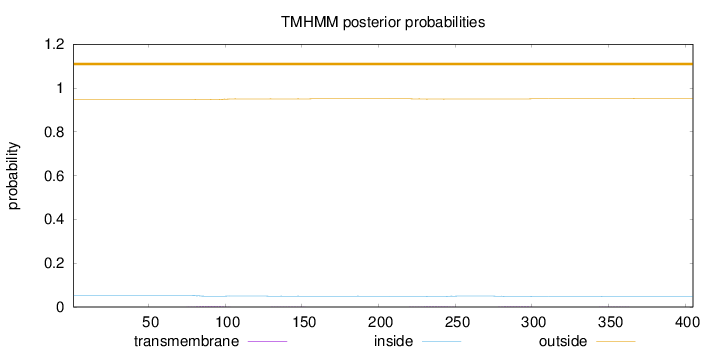
Topology
Length:
405
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.14177
Exp number, first 60 AAs:
0
Total prob of N-in:
0.05286
outside
1 - 405
Population Genetic Test Statistics
Pi
292.31582
Theta
173.696906
Tajima's D
1.818438
CLR
0.05395
CSRT
0.853557322133893
Interpretation
Uncertain
