Gene
KWMTBOMO15144
Pre Gene Modal
BGIBMGA004926
Annotation
PREDICTED:_cysteine_protease_ATG4D_[Bombyx_mori]
Full name
Cysteine protease
+ More
Cysteine protease ATG4D
Cysteine protease ATG4D
Alternative Name
AUT-like 4 cysteine endopeptidase
Autophagin-4
Autophagy-related cysteine endopeptidase 4
Autophagy-related protein 4 homolog D
Autophagin-4
Autophagy-related cysteine endopeptidase 4
Autophagy-related protein 4 homolog D
Location in the cell
Cytoplasmic Reliability : 1.394 Extracellular Reliability : 1.012
Sequence
CDS
ATGGAAGCAACTGCACATCAACCAATGGAACAAATATATGGAGAGGGTATAGAAGGATTTAAATCAGATTTTGTAAGTAAAATATGGATGACTTACAGAAGGGAATTCCCTACTATGACAGGTTCAACTTTTACAACTGATTGTGGTTGGGGTTGTATGCTTCGAAGTGGACAAATGATGTTGGCTCAGGCTTTAGTTTGCCATTTCCTAGGCCGTTCATGGAGATGGTTACCTGAGAAACCCATTCAAAATGCTAGAGAATTTCAAGAAGATTGTTTACATAGAAAAATTATAAAGTGGTTTGGAGATAAGTCTTCAGTTAATAGTCCTCTGTCAATACATCAGATGGTGAGTTTGGGAGAAGCTTTAGGCAAGAAGCCAGGTGATTGGTATGGCCCTGCATCTGTAGCACATTGTTTGAAATCTCTAATTGCATCAGCTTCAAAAGAGAATTATGAATTTGATCATTTAGAAGTTTATGTTGCTCAAGATTCAACTGTTTATATTCAAGATATATATTCTATGTGTCAGTTGTTACATGGAGCTTGGAAGTCACTTATTCTTCTAGTTCCAGTGAAATTAGGCACTGAAAAATTTAATCCAATATATGGCCCATGCCTCACATCATTATTAACATTAGATTTTTGCATTGGAATCATTGGTGGAAGGCCAAAACATTCTCTATATTTTGTTGGCTATCAAGATGACAAACTCATTCATCTGGATCCACATTACTGCCAAGAAATGGTAGATGTTTGGCAACCCAATTTCTCTTTACAAAGTTTTCATTGTCGTTCTCCAAGAAAAATGCCACTTGCAAAGATGGATCCATCATGTTGTATTGGTTTTTATCTTGGAACCCAACATGATTTTGAAACATTTATAAATATAATAAAATCATTCCTAGTACCTCAGGGTGTATCATCAGGATATGAATATCCTATATTTACTCTAAATAATGGCTCCTGTGATTCACTTATGGATACTCCTAATGCTAGATATTCCTTTTTTGAAACAGAACAACATTGGGTCACTCCACATATGCAAGACAGCGATACAGACTTAGAGTCTGAAGAATTTGTTTTAGTTTAG
Protein
MEATAHQPMEQIYGEGIEGFKSDFVSKIWMTYRREFPTMTGSTFTTDCGWGCMLRSGQMMLAQALVCHFLGRSWRWLPEKPIQNAREFQEDCLHRKIIKWFGDKSSVNSPLSIHQMVSLGEALGKKPGDWYGPASVAHCLKSLIASASKENYEFDHLEVYVAQDSTVYIQDIYSMCQLLHGAWKSLILLVPVKLGTEKFNPIYGPCLTSLLTLDFCIGIIGGRPKHSLYFVGYQDDKLIHLDPHYCQEMVDVWQPNFSLQSFHCRSPRKMPLAKMDPSCCIGFYLGTQHDFETFINIIKSFLVPQGVSSGYEYPIFTLNNGSCDSLMDTPNARYSFFETEQHWVTPHMQDSDTDLESEEFVLV
Summary
Description
Cysteine protease required for the cytoplasm to vacuole transport (Cvt) and autophagy.
Cysteine protease ATG4D: Cysteine protease required for the cytoplasm to vacuole transport (Cvt) and autophagy. Cleaves the C-terminal amino acid of ATG8 family proteins MAP1LC3 and GABARAPL2, to reveal a C-terminal glycine. Exposure of the glycine at the C-terminus is essential for ATG8 proteins conjugation to phosphatidylethanolamine (PE) and insertion to membranes, which is necessary for autophagy. Has also an activity of delipidating enzyme for the PE-conjugated forms.
Cysteine protease ATG4D, mitochondrial: Plays a role as an autophagy regulator that links mitochondrial dysfunction with apoptosis. The mitochondrial import of ATG4D during cellular stress and differentiation may play important roles in the regulation of mitochondrial physiology, ROS, mitophagy and cell viability (By similarity).
Cysteine protease ATG4D: Cysteine protease required for the cytoplasm to vacuole transport (Cvt) and autophagy. Cleaves the C-terminal amino acid of ATG8 family proteins MAP1LC3 and GABARAPL2, to reveal a C-terminal glycine. Exposure of the glycine at the C-terminus is essential for ATG8 proteins conjugation to phosphatidylethanolamine (PE) and insertion to membranes, which is necessary for autophagy. Has also an activity of delipidating enzyme for the PE-conjugated forms.
Cysteine protease ATG4D, mitochondrial: Plays a role as an autophagy regulator that links mitochondrial dysfunction with apoptosis. The mitochondrial import of ATG4D during cellular stress and differentiation may play important roles in the regulation of mitochondrial physiology, ROS, mitophagy and cell viability (By similarity).
Similarity
Belongs to the peptidase C54 family.
Keywords
Apoptosis
Autophagy
Complete proteome
Cytoplasm
Direct protein sequencing
Hydrolase
Mitochondrion
Phosphoprotein
Protease
Protein transport
Reference proteome
Thiol protease
Transport
Ubl conjugation pathway
Feature
chain Cysteine protease ATG4D
Uniprot
H9J5Y5
B7TYK3
A0A2A4JNP6
A0A0L7KT31
A0A2H1WZJ1
A0A212ETV7
+ More
A0A194RK62 A0A1E1WIK0 A0A2J7PPA3 A0A2J7PP70 A0A2J7PP62 A0A067RD38 A0A1Y1M762 A0A026X3C0 D6W716 E2AN45 A0A139WNQ2 N6UDY1 A0A1B0FEE9 E2BJM9 A0A088A1U1 U4UWJ6 A0A1A9YSV2 B4KAQ1 A0A0Q9X9V0 A0A1B0B2Q8 A0A1A9ZZ01 B4JU42 A0A034WKX6 W8BYD8 W8C9F4 A0A1A9VFZ3 A0A0K8VFZ3 A0A0K8V823 A0A1I8QE56 A0A1I8QE05 A0A154P6E0 A0A1I8M0T4 A0A0L0BN65 A0A1W4VV08 F4WFG7 A0A158NSA5 A0A195BSF8 A0A0K8TL78 A0A0R1E3F3 B4PS40 A0A0L7QU64 B4M5Z5 A0A151JLE5 B3P0V8 A0A0Q9WDD0 A0A151JYF0 B4HM23 A0A3B0JK89 Q9VF80 B4QYI4 A0A3B0K6W7 Q8T0A2 B4NAU0 A0A0P8Y2J7 B3MSU1 A0A0R3NF26 A0A0M4EVS4 B4GNU0 Q29BE2 A0A336M5E9 A0A195CX72 A0A336L0T6 A0A151XK73 A0A1L8E515 A0A0N8B4Y2 A0A0P5S5Q6 A0A0P6DWW8 A0A0P5JXL2 A0A0P5ESD2 E9GBM8 A0A2A3ENI3 E9J4X0 A0A0P6B279 A0A1S3KAA7 A0A131XYR8 A0A2R5LM58 A0A1E1XKI8 A0A1Z5LD75 T1J1K5 A0A1E1XG87 A0A131XHZ8 G1T6V0 Q8BGV9 A0A0S2ZXR5 Q3U4N6 A0A293N4W1 A0A3B3UQ71 D2HGY4 A0A087Y4F0 A0A3B3WX25 A0A3Q2CAZ0 G1LY72
A0A194RK62 A0A1E1WIK0 A0A2J7PPA3 A0A2J7PP70 A0A2J7PP62 A0A067RD38 A0A1Y1M762 A0A026X3C0 D6W716 E2AN45 A0A139WNQ2 N6UDY1 A0A1B0FEE9 E2BJM9 A0A088A1U1 U4UWJ6 A0A1A9YSV2 B4KAQ1 A0A0Q9X9V0 A0A1B0B2Q8 A0A1A9ZZ01 B4JU42 A0A034WKX6 W8BYD8 W8C9F4 A0A1A9VFZ3 A0A0K8VFZ3 A0A0K8V823 A0A1I8QE56 A0A1I8QE05 A0A154P6E0 A0A1I8M0T4 A0A0L0BN65 A0A1W4VV08 F4WFG7 A0A158NSA5 A0A195BSF8 A0A0K8TL78 A0A0R1E3F3 B4PS40 A0A0L7QU64 B4M5Z5 A0A151JLE5 B3P0V8 A0A0Q9WDD0 A0A151JYF0 B4HM23 A0A3B0JK89 Q9VF80 B4QYI4 A0A3B0K6W7 Q8T0A2 B4NAU0 A0A0P8Y2J7 B3MSU1 A0A0R3NF26 A0A0M4EVS4 B4GNU0 Q29BE2 A0A336M5E9 A0A195CX72 A0A336L0T6 A0A151XK73 A0A1L8E515 A0A0N8B4Y2 A0A0P5S5Q6 A0A0P6DWW8 A0A0P5JXL2 A0A0P5ESD2 E9GBM8 A0A2A3ENI3 E9J4X0 A0A0P6B279 A0A1S3KAA7 A0A131XYR8 A0A2R5LM58 A0A1E1XKI8 A0A1Z5LD75 T1J1K5 A0A1E1XG87 A0A131XHZ8 G1T6V0 Q8BGV9 A0A0S2ZXR5 Q3U4N6 A0A293N4W1 A0A3B3UQ71 D2HGY4 A0A087Y4F0 A0A3B3WX25 A0A3Q2CAZ0 G1LY72
EC Number
3.4.22.-
Pubmed
19121390
19470186
26227816
22118469
26354079
24845553
+ More
28004739 24508170 30249741 18362917 19820115 20798317 23537049 17994087 25348373 24495485 25315136 26108605 21719571 21347285 26369729 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 21292972 21282665 29652888 29209593 28528879 28503490 28049606 21993624 12446702 16141072 15489334 10349636 11042159 11076861 11217851 12466851 16141073 20010809
28004739 24508170 30249741 18362917 19820115 20798317 23537049 17994087 25348373 24495485 25315136 26108605 21719571 21347285 26369729 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 21292972 21282665 29652888 29209593 28528879 28503490 28049606 21993624 12446702 16141072 15489334 10349636 11042159 11076861 11217851 12466851 16141073 20010809
EMBL
BABH01029969
FJ416326
ACJ46060.1
NWSH01001030
PCG73042.1
JTDY01006181
+ More
KOB66211.1 ODYU01012229 SOQ58427.1 AGBW02012531 OWR44884.1 KQ460045 KPJ18223.1 GDQN01004211 JAT86843.1 NEVH01023279 PNF18136.1 PNF18135.1 PNF18134.1 KK852539 KDR21791.1 GEZM01042602 JAV79766.1 KK107019 QOIP01000007 EZA62757.1 RLU20531.1 KQ971307 EFA11495.1 GL441072 EFN65142.1 KYB29638.1 APGK01039147 KB740967 ENN76867.1 CCAG010001986 GL448604 EFN84087.1 KB632399 ERL94645.1 CH933806 EDW16788.2 KRG02298.1 JXJN01007687 CH916374 EDV91012.1 GAKP01004515 GAKP01004514 GAKP01004512 JAC54438.1 GAMC01002198 JAC04358.1 GAMC01002200 GAMC01002199 JAC04357.1 GDHF01014839 JAI37475.1 GDHF01027672 GDHF01017411 JAI24642.1 JAI34903.1 KQ434819 KZC06914.1 JRES01001623 KNC21452.1 GL888116 EGI67039.1 ADTU01024688 KQ976421 KYM89088.1 GDAI01002506 JAI15097.1 CM000160 KRK03768.1 EDW97460.1 KQ414735 KOC62160.1 CH940652 EDW59071.2 KQ979011 KYN26629.1 CH954181 EDV49006.1 KRF78661.1 KQ981486 KYN41427.1 CH480815 EDW42060.1 OUUW01000005 SPP80742.1 AE014297 BT100116 AAF55180.2 ACX83596.1 CM000364 EDX12829.1 SPP80741.1 AY069453 AAL39598.1 CH964232 EDW80904.2 CH902623 KPU72801.1 EDV30331.2 CM000070 KRS99747.1 CP012526 ALC47492.1 CH479186 EDW38823.1 EAL27056.4 UFQT01000415 SSX24099.1 KQ977171 KYN05152.1 UFQS01001255 UFQT01001255 SSX09960.1 SSX29683.1 KQ982045 KYQ60731.1 GFDF01000257 JAV13827.1 GDIQ01212403 JAK39322.1 GDIQ01109182 LRGB01002860 JAL42544.1 KZS05873.1 GDIQ01074906 JAN19831.1 GDIQ01192092 JAK59633.1 GDIP01144037 JAJ79365.1 GL732538 EFX82987.1 KZ288215 PBC32699.1 GL768121 EFZ12136.1 GDIP01021217 JAM82498.1 GEFM01003647 GEGO01001913 JAP72149.1 JAR93491.1 GGLE01006468 MBY10594.1 GFAA01003602 JAT99832.1 GFJQ02001695 JAW05275.1 JH431789 GFAC01000960 JAT98228.1 GEFH01002746 JAP65835.1 AJ312333 AK036571 AK081605 AK081002 BC030861 BC069851 KT194093 ALQ43549.1 AK154130 BAE32395.1 GFWV01023069 MAA47796.1 GL192824 EFB12981.1 AYCK01004499 ACTA01058381 ACTA01066381
KOB66211.1 ODYU01012229 SOQ58427.1 AGBW02012531 OWR44884.1 KQ460045 KPJ18223.1 GDQN01004211 JAT86843.1 NEVH01023279 PNF18136.1 PNF18135.1 PNF18134.1 KK852539 KDR21791.1 GEZM01042602 JAV79766.1 KK107019 QOIP01000007 EZA62757.1 RLU20531.1 KQ971307 EFA11495.1 GL441072 EFN65142.1 KYB29638.1 APGK01039147 KB740967 ENN76867.1 CCAG010001986 GL448604 EFN84087.1 KB632399 ERL94645.1 CH933806 EDW16788.2 KRG02298.1 JXJN01007687 CH916374 EDV91012.1 GAKP01004515 GAKP01004514 GAKP01004512 JAC54438.1 GAMC01002198 JAC04358.1 GAMC01002200 GAMC01002199 JAC04357.1 GDHF01014839 JAI37475.1 GDHF01027672 GDHF01017411 JAI24642.1 JAI34903.1 KQ434819 KZC06914.1 JRES01001623 KNC21452.1 GL888116 EGI67039.1 ADTU01024688 KQ976421 KYM89088.1 GDAI01002506 JAI15097.1 CM000160 KRK03768.1 EDW97460.1 KQ414735 KOC62160.1 CH940652 EDW59071.2 KQ979011 KYN26629.1 CH954181 EDV49006.1 KRF78661.1 KQ981486 KYN41427.1 CH480815 EDW42060.1 OUUW01000005 SPP80742.1 AE014297 BT100116 AAF55180.2 ACX83596.1 CM000364 EDX12829.1 SPP80741.1 AY069453 AAL39598.1 CH964232 EDW80904.2 CH902623 KPU72801.1 EDV30331.2 CM000070 KRS99747.1 CP012526 ALC47492.1 CH479186 EDW38823.1 EAL27056.4 UFQT01000415 SSX24099.1 KQ977171 KYN05152.1 UFQS01001255 UFQT01001255 SSX09960.1 SSX29683.1 KQ982045 KYQ60731.1 GFDF01000257 JAV13827.1 GDIQ01212403 JAK39322.1 GDIQ01109182 LRGB01002860 JAL42544.1 KZS05873.1 GDIQ01074906 JAN19831.1 GDIQ01192092 JAK59633.1 GDIP01144037 JAJ79365.1 GL732538 EFX82987.1 KZ288215 PBC32699.1 GL768121 EFZ12136.1 GDIP01021217 JAM82498.1 GEFM01003647 GEGO01001913 JAP72149.1 JAR93491.1 GGLE01006468 MBY10594.1 GFAA01003602 JAT99832.1 GFJQ02001695 JAW05275.1 JH431789 GFAC01000960 JAT98228.1 GEFH01002746 JAP65835.1 AJ312333 AK036571 AK081605 AK081002 BC030861 BC069851 KT194093 ALQ43549.1 AK154130 BAE32395.1 GFWV01023069 MAA47796.1 GL192824 EFB12981.1 AYCK01004499 ACTA01058381 ACTA01066381
Proteomes
UP000005204
UP000218220
UP000037510
UP000007151
UP000053240
UP000235965
+ More
UP000027135 UP000053097 UP000279307 UP000007266 UP000000311 UP000019118 UP000092444 UP000008237 UP000005203 UP000030742 UP000092443 UP000009192 UP000092460 UP000092445 UP000001070 UP000078200 UP000095300 UP000076502 UP000095301 UP000037069 UP000192221 UP000007755 UP000005205 UP000078540 UP000002282 UP000053825 UP000008792 UP000078492 UP000008711 UP000078541 UP000001292 UP000268350 UP000000803 UP000000304 UP000007798 UP000007801 UP000001819 UP000092553 UP000008744 UP000078542 UP000075809 UP000076858 UP000000305 UP000242457 UP000085678 UP000001811 UP000000589 UP000261500 UP000028760 UP000261480 UP000265020 UP000008912
UP000027135 UP000053097 UP000279307 UP000007266 UP000000311 UP000019118 UP000092444 UP000008237 UP000005203 UP000030742 UP000092443 UP000009192 UP000092460 UP000092445 UP000001070 UP000078200 UP000095300 UP000076502 UP000095301 UP000037069 UP000192221 UP000007755 UP000005205 UP000078540 UP000002282 UP000053825 UP000008792 UP000078492 UP000008711 UP000078541 UP000001292 UP000268350 UP000000803 UP000000304 UP000007798 UP000007801 UP000001819 UP000092553 UP000008744 UP000078542 UP000075809 UP000076858 UP000000305 UP000242457 UP000085678 UP000001811 UP000000589 UP000261500 UP000028760 UP000261480 UP000265020 UP000008912
Pfam
PF03416 Peptidase_C54
SUPFAM
SSF54001
SSF54001
ProteinModelPortal
H9J5Y5
B7TYK3
A0A2A4JNP6
A0A0L7KT31
A0A2H1WZJ1
A0A212ETV7
+ More
A0A194RK62 A0A1E1WIK0 A0A2J7PPA3 A0A2J7PP70 A0A2J7PP62 A0A067RD38 A0A1Y1M762 A0A026X3C0 D6W716 E2AN45 A0A139WNQ2 N6UDY1 A0A1B0FEE9 E2BJM9 A0A088A1U1 U4UWJ6 A0A1A9YSV2 B4KAQ1 A0A0Q9X9V0 A0A1B0B2Q8 A0A1A9ZZ01 B4JU42 A0A034WKX6 W8BYD8 W8C9F4 A0A1A9VFZ3 A0A0K8VFZ3 A0A0K8V823 A0A1I8QE56 A0A1I8QE05 A0A154P6E0 A0A1I8M0T4 A0A0L0BN65 A0A1W4VV08 F4WFG7 A0A158NSA5 A0A195BSF8 A0A0K8TL78 A0A0R1E3F3 B4PS40 A0A0L7QU64 B4M5Z5 A0A151JLE5 B3P0V8 A0A0Q9WDD0 A0A151JYF0 B4HM23 A0A3B0JK89 Q9VF80 B4QYI4 A0A3B0K6W7 Q8T0A2 B4NAU0 A0A0P8Y2J7 B3MSU1 A0A0R3NF26 A0A0M4EVS4 B4GNU0 Q29BE2 A0A336M5E9 A0A195CX72 A0A336L0T6 A0A151XK73 A0A1L8E515 A0A0N8B4Y2 A0A0P5S5Q6 A0A0P6DWW8 A0A0P5JXL2 A0A0P5ESD2 E9GBM8 A0A2A3ENI3 E9J4X0 A0A0P6B279 A0A1S3KAA7 A0A131XYR8 A0A2R5LM58 A0A1E1XKI8 A0A1Z5LD75 T1J1K5 A0A1E1XG87 A0A131XHZ8 G1T6V0 Q8BGV9 A0A0S2ZXR5 Q3U4N6 A0A293N4W1 A0A3B3UQ71 D2HGY4 A0A087Y4F0 A0A3B3WX25 A0A3Q2CAZ0 G1LY72
A0A194RK62 A0A1E1WIK0 A0A2J7PPA3 A0A2J7PP70 A0A2J7PP62 A0A067RD38 A0A1Y1M762 A0A026X3C0 D6W716 E2AN45 A0A139WNQ2 N6UDY1 A0A1B0FEE9 E2BJM9 A0A088A1U1 U4UWJ6 A0A1A9YSV2 B4KAQ1 A0A0Q9X9V0 A0A1B0B2Q8 A0A1A9ZZ01 B4JU42 A0A034WKX6 W8BYD8 W8C9F4 A0A1A9VFZ3 A0A0K8VFZ3 A0A0K8V823 A0A1I8QE56 A0A1I8QE05 A0A154P6E0 A0A1I8M0T4 A0A0L0BN65 A0A1W4VV08 F4WFG7 A0A158NSA5 A0A195BSF8 A0A0K8TL78 A0A0R1E3F3 B4PS40 A0A0L7QU64 B4M5Z5 A0A151JLE5 B3P0V8 A0A0Q9WDD0 A0A151JYF0 B4HM23 A0A3B0JK89 Q9VF80 B4QYI4 A0A3B0K6W7 Q8T0A2 B4NAU0 A0A0P8Y2J7 B3MSU1 A0A0R3NF26 A0A0M4EVS4 B4GNU0 Q29BE2 A0A336M5E9 A0A195CX72 A0A336L0T6 A0A151XK73 A0A1L8E515 A0A0N8B4Y2 A0A0P5S5Q6 A0A0P6DWW8 A0A0P5JXL2 A0A0P5ESD2 E9GBM8 A0A2A3ENI3 E9J4X0 A0A0P6B279 A0A1S3KAA7 A0A131XYR8 A0A2R5LM58 A0A1E1XKI8 A0A1Z5LD75 T1J1K5 A0A1E1XG87 A0A131XHZ8 G1T6V0 Q8BGV9 A0A0S2ZXR5 Q3U4N6 A0A293N4W1 A0A3B3UQ71 D2HGY4 A0A087Y4F0 A0A3B3WX25 A0A3Q2CAZ0 G1LY72
PDB
2P82
E-value=7.47139e-55,
Score=541
Ontologies
PATHWAY
GO
PANTHER
Topology
Subcellular location
Cytoplasm
Mitochondrion matrix Imported into mitochondrial matrix after cleavage by CASP3 during oxidative stress and cell death. With evidence from 2 publications.
Mitochondrion matrix Imported into mitochondrial matrix after cleavage by CASP3 during oxidative stress and cell death. With evidence from 2 publications.
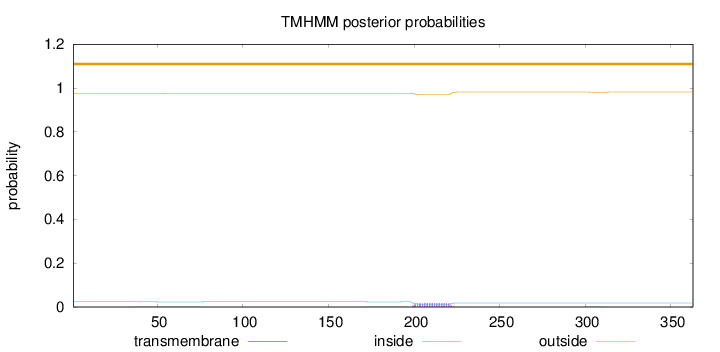
Length:
363
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.42238
Exp number, first 60 AAs:
0.01797
Total prob of N-in:
0.02413
outside
1 - 363
Population Genetic Test Statistics
Pi
297.996658
Theta
215.176641
Tajima's D
0.969729
CLR
0.016304
CSRT
0.652767361631918
Interpretation
Uncertain
