Gene
KWMTBOMO15140
Pre Gene Modal
BGIBMGA004923
Annotation
PREDICTED:_luciferin_4-monooxygenase-like_[Bombyx_mori]
Full name
Luciferin 4-monooxygenase
Location in the cell
Cytoplasmic Reliability : 3.396
Sequence
CDS
ATGCCCAAAGTCGTTGACATTAATACTCGACAAACATTAGGATCGTATGAACGAGGTGAAGTCTGCGTTAAAGGACCCCTGGTGATGAAAGGCTACATGAATAACGATCAAGCTAATGAAGATATAAAGGACGATGAGGGTTTCCTCAAAACTGGAGATATCGGATATTATGATTCAGACGGCTGTTTCTACATAGTCGACAGACTGAAAGAACTTATCAAATACAAAGGACATCAGGTAGCTCCAGCAATGATAGAGCGCGTACTGCTGCAGCACAGCGACGTGTTGGAGTGCGGAGTGGTGGGAGCGCCCGACGAGTTGGCTGGCGAGCTGCCCACCGCCTTCGTAGTGCCCAAGCCTGACGTCACGCTCACAGAAAGAGAACTGCTAGACTTCACTAATGAACGGTTATCCCCAATCACGCGAATACGAGGAGGTATATTCTTCGTGGGGGAAATCCCAAAGAATGCAACCGGAAAAATACTACGAAAAGTACTTAAAGCTAATTTAATAGCAAATAAATGA
Protein
MPKVVDINTRQTLGSYERGEVCVKGPLVMKGYMNNDQANEDIKDDEGFLKTGDIGYYDSDGCFYIVDRLKELIKYKGHQVAPAMIERVLLQHSDVLECGVVGAPDELAGELPTAFVVPKPDVTLTERELLDFTNERLSPITRIRGGIFFVGEIPKNATGKILRKVLKANLIANK
Summary
Description
Produces green light.
Catalytic Activity
ATP + firefly D-luciferin + O2 = AMP + CO2 + diphosphate + firefly oxyluciferin + hnu
Cofactor
Mg(2+)
Similarity
Belongs to the ATP-dependent AMP-binding enzyme family.
Keywords
ATP-binding
Luminescence
Magnesium
Metal-binding
Monooxygenase
Nucleotide-binding
Oxidoreductase
Peroxisome
Photoprotein
Feature
chain Luciferin 4-monooxygenase
Uniprot
H9J5Y2
H9J5Y0
A0A194PHR8
A0A2H1W987
A0A194RK67
A0A2A4JN67
+ More
A0A2A4JMM7 H9J5X9 A0A3S2NFX7 H9J647 A0A194PGU6 A0A182MLJ2 A0A2H1WNI5 A0A2A4JNM0 A0A084VAF1 A0A194PT23 A0A182JAX9 A0A3S2PFN8 A0A194RLN8 A0A1I8JUI2 H9J2F6 G8GE17 A0A2C9GUN5 Q05KC0 A0A212ETT6 A0A194QZM0 A0A182Q9Y0 A0A0L7KQP2 A0A084W803 I4DPZ6 A0A1I8JVG1 A0A0P4Z3R1 A0A0P5EQ02 A0A1Y9H2Q7 D2DHG8 A0A1Y9IUV9 A0A2A4J232 A0A240PK82 A0A194PTD5 A0A240PPP9 A0A1I8JTD3 A0A0P5L0B8 A0A182F8R8 Q1WLP6 A0A2M4CMC1 A0A1Y9IV15 A0A0P5G2W6 A0A0P5T6B2 Q5TVT3 Q2ACC9 W5JDW2 E3SH50 A0A2J7QLT8 A0A2M4CH72 A0A1I8JUZ8 W5JL40 A0A194PUC3 A0A067R342 A0A0P5U9Q6 A0A240PLR9 A0A182X4T0 A0A2B4RUS4 A0A2M4BKV7 A0A1I8JSN8 A0A1I8JUE9 A0A182TA88 A0A1Y9H2R4 A0A182YM73 A0A0P5WBB6 A0A2M4BJ94 W5JFE9 A0A2J7QLX8 Q7Q503 A0A182I9E2 A0A0P5ZDU6 A0A182LQ72 A0A0N8E2B3 Q2ABY2 A0A1S3HMF6 A0A0N8D2I0 A0A1I8JVV1 Q01158 A0A1Q2TT53 H1AD94 A0A0N8ADN0 A0A182Q5M2 A0A0P4ZJZ9 A0A2M3Z9G4 A0A0T6B4U7 A0A0B7BDP1 A0A0P5EAE4 A0A1I8JVD9 Q8T6U3 A0A2M4BJS1 Q8ITG5 Q8I0E8 A0A0P6BFI2 Q8ITG4 A0A0P5IPP8 A0A2M4AK32
A0A2A4JMM7 H9J5X9 A0A3S2NFX7 H9J647 A0A194PGU6 A0A182MLJ2 A0A2H1WNI5 A0A2A4JNM0 A0A084VAF1 A0A194PT23 A0A182JAX9 A0A3S2PFN8 A0A194RLN8 A0A1I8JUI2 H9J2F6 G8GE17 A0A2C9GUN5 Q05KC0 A0A212ETT6 A0A194QZM0 A0A182Q9Y0 A0A0L7KQP2 A0A084W803 I4DPZ6 A0A1I8JVG1 A0A0P4Z3R1 A0A0P5EQ02 A0A1Y9H2Q7 D2DHG8 A0A1Y9IUV9 A0A2A4J232 A0A240PK82 A0A194PTD5 A0A240PPP9 A0A1I8JTD3 A0A0P5L0B8 A0A182F8R8 Q1WLP6 A0A2M4CMC1 A0A1Y9IV15 A0A0P5G2W6 A0A0P5T6B2 Q5TVT3 Q2ACC9 W5JDW2 E3SH50 A0A2J7QLT8 A0A2M4CH72 A0A1I8JUZ8 W5JL40 A0A194PUC3 A0A067R342 A0A0P5U9Q6 A0A240PLR9 A0A182X4T0 A0A2B4RUS4 A0A2M4BKV7 A0A1I8JSN8 A0A1I8JUE9 A0A182TA88 A0A1Y9H2R4 A0A182YM73 A0A0P5WBB6 A0A2M4BJ94 W5JFE9 A0A2J7QLX8 Q7Q503 A0A182I9E2 A0A0P5ZDU6 A0A182LQ72 A0A0N8E2B3 Q2ABY2 A0A1S3HMF6 A0A0N8D2I0 A0A1I8JVV1 Q01158 A0A1Q2TT53 H1AD94 A0A0N8ADN0 A0A182Q5M2 A0A0P4ZJZ9 A0A2M3Z9G4 A0A0T6B4U7 A0A0B7BDP1 A0A0P5EAE4 A0A1I8JVD9 Q8T6U3 A0A2M4BJS1 Q8ITG5 Q8I0E8 A0A0P6BFI2 Q8ITG4 A0A0P5IPP8 A0A2M4AK32
EC Number
1.13.12.7
Pubmed
EMBL
BABH01029974
BABH01029976
KQ459604
KPI92264.1
ODYU01007142
SOQ49659.1
+ More
KQ460045 KPJ18228.1 NWSH01001030 PCG73034.1 PCG73036.1 BABH01029979 BABH01029980 RSAL01000053 RVE50075.1 BABH01029982 KPI92263.1 AXCM01004086 ODYU01009511 SOQ54004.1 PCG73033.1 ATLV01003206 KE524124 KFB34945.1 KQ459593 KPI96566.1 RVE50074.1 KPJ18230.1 BABH01007891 JN127350 AET05796.1 AB255748 BAF34360.1 AGBW02012531 OWR44889.1 KQ460890 KPJ10988.1 AXCN02000090 JTDY01007153 KOB65430.1 ATLV01021336 KE525316 KFB46347.1 AK403861 BAM19986.1 GDIP01230533 JAI92868.1 GDIP01156704 JAJ66698.1 FJ545728 ACT68596.1 NWSH01003686 PCG65919.1 KPI96567.1 APCN01002222 GDIQ01184560 JAK67165.1 DQ137139 AAZ74651.1 GGFL01002163 MBW66341.1 GDIQ01247095 JAK04630.1 GDIP01136942 JAL66772.1 AAAB01008820 EAL41501.4 AB196455 BAE80728.1 ADMH02001657 ETN61533.1 GU062703 ADK56478.1 NEVH01013241 PNF29543.1 GGFL01000090 MBW64268.1 ADMH02001214 ETN63630.1 KPI96573.1 KK852730 KDR17523.1 GDIP01117721 JAL85993.1 LSMT01000312 PFX20569.1 GGFJ01004568 MBW53709.1 GDIP01088499 GDIP01088498 JAM15217.1 GGFJ01003984 MBW53125.1 ETN61534.1 PNF29548.1 AAAB01008961 EAA11995.3 APCN01008266 GDIP01046867 LRGB01002371 JAM56848.1 KZS08045.1 GDIQ01068764 JAN25973.1 AB220162 BAE80731.1 GDIP01075207 JAM28508.1 X66919 AB909130 BAW94791.1 AB644226 BAL46510.1 GDIP01157697 JAJ65705.1 GDIP01211436 JAJ11966.1 GGFM01004391 MBW25142.1 LJIG01009801 KRT82354.1 HACG01044107 CEK90972.1 GDIP01144886 JAJ78516.1 AF420006 AAM00429.1 GGFJ01004113 MBW53254.1 AF486800 AAN40975.1 AF486802 AF486803 AAN40977.1 GDIP01015371 JAM88344.1 AF486801 AAN40976.1 GDIQ01210454 JAK41271.1 GGFK01007825 MBW41146.1
KQ460045 KPJ18228.1 NWSH01001030 PCG73034.1 PCG73036.1 BABH01029979 BABH01029980 RSAL01000053 RVE50075.1 BABH01029982 KPI92263.1 AXCM01004086 ODYU01009511 SOQ54004.1 PCG73033.1 ATLV01003206 KE524124 KFB34945.1 KQ459593 KPI96566.1 RVE50074.1 KPJ18230.1 BABH01007891 JN127350 AET05796.1 AB255748 BAF34360.1 AGBW02012531 OWR44889.1 KQ460890 KPJ10988.1 AXCN02000090 JTDY01007153 KOB65430.1 ATLV01021336 KE525316 KFB46347.1 AK403861 BAM19986.1 GDIP01230533 JAI92868.1 GDIP01156704 JAJ66698.1 FJ545728 ACT68596.1 NWSH01003686 PCG65919.1 KPI96567.1 APCN01002222 GDIQ01184560 JAK67165.1 DQ137139 AAZ74651.1 GGFL01002163 MBW66341.1 GDIQ01247095 JAK04630.1 GDIP01136942 JAL66772.1 AAAB01008820 EAL41501.4 AB196455 BAE80728.1 ADMH02001657 ETN61533.1 GU062703 ADK56478.1 NEVH01013241 PNF29543.1 GGFL01000090 MBW64268.1 ADMH02001214 ETN63630.1 KPI96573.1 KK852730 KDR17523.1 GDIP01117721 JAL85993.1 LSMT01000312 PFX20569.1 GGFJ01004568 MBW53709.1 GDIP01088499 GDIP01088498 JAM15217.1 GGFJ01003984 MBW53125.1 ETN61534.1 PNF29548.1 AAAB01008961 EAA11995.3 APCN01008266 GDIP01046867 LRGB01002371 JAM56848.1 KZS08045.1 GDIQ01068764 JAN25973.1 AB220162 BAE80731.1 GDIP01075207 JAM28508.1 X66919 AB909130 BAW94791.1 AB644226 BAL46510.1 GDIP01157697 JAJ65705.1 GDIP01211436 JAJ11966.1 GGFM01004391 MBW25142.1 LJIG01009801 KRT82354.1 HACG01044107 CEK90972.1 GDIP01144886 JAJ78516.1 AF420006 AAM00429.1 GGFJ01004113 MBW53254.1 AF486800 AAN40975.1 AF486802 AF486803 AAN40977.1 GDIP01015371 JAM88344.1 AF486801 AAN40976.1 GDIQ01210454 JAK41271.1 GGFK01007825 MBW41146.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000218220
UP000283053
UP000075883
+ More
UP000030765 UP000075880 UP000075900 UP000007151 UP000075886 UP000037510 UP000075903 UP000075884 UP000075920 UP000075885 UP000075840 UP000069272 UP000007062 UP000000673 UP000235965 UP000075902 UP000027135 UP000075881 UP000076407 UP000225706 UP000075901 UP000076408 UP000076858 UP000075882 UP000085678
UP000030765 UP000075880 UP000075900 UP000007151 UP000075886 UP000037510 UP000075903 UP000075884 UP000075920 UP000075885 UP000075840 UP000069272 UP000007062 UP000000673 UP000235965 UP000075902 UP000027135 UP000075881 UP000076407 UP000225706 UP000075901 UP000076408 UP000076858 UP000075882 UP000085678
Interpro
Gene 3D
ProteinModelPortal
H9J5Y2
H9J5Y0
A0A194PHR8
A0A2H1W987
A0A194RK67
A0A2A4JN67
+ More
A0A2A4JMM7 H9J5X9 A0A3S2NFX7 H9J647 A0A194PGU6 A0A182MLJ2 A0A2H1WNI5 A0A2A4JNM0 A0A084VAF1 A0A194PT23 A0A182JAX9 A0A3S2PFN8 A0A194RLN8 A0A1I8JUI2 H9J2F6 G8GE17 A0A2C9GUN5 Q05KC0 A0A212ETT6 A0A194QZM0 A0A182Q9Y0 A0A0L7KQP2 A0A084W803 I4DPZ6 A0A1I8JVG1 A0A0P4Z3R1 A0A0P5EQ02 A0A1Y9H2Q7 D2DHG8 A0A1Y9IUV9 A0A2A4J232 A0A240PK82 A0A194PTD5 A0A240PPP9 A0A1I8JTD3 A0A0P5L0B8 A0A182F8R8 Q1WLP6 A0A2M4CMC1 A0A1Y9IV15 A0A0P5G2W6 A0A0P5T6B2 Q5TVT3 Q2ACC9 W5JDW2 E3SH50 A0A2J7QLT8 A0A2M4CH72 A0A1I8JUZ8 W5JL40 A0A194PUC3 A0A067R342 A0A0P5U9Q6 A0A240PLR9 A0A182X4T0 A0A2B4RUS4 A0A2M4BKV7 A0A1I8JSN8 A0A1I8JUE9 A0A182TA88 A0A1Y9H2R4 A0A182YM73 A0A0P5WBB6 A0A2M4BJ94 W5JFE9 A0A2J7QLX8 Q7Q503 A0A182I9E2 A0A0P5ZDU6 A0A182LQ72 A0A0N8E2B3 Q2ABY2 A0A1S3HMF6 A0A0N8D2I0 A0A1I8JVV1 Q01158 A0A1Q2TT53 H1AD94 A0A0N8ADN0 A0A182Q5M2 A0A0P4ZJZ9 A0A2M3Z9G4 A0A0T6B4U7 A0A0B7BDP1 A0A0P5EAE4 A0A1I8JVD9 Q8T6U3 A0A2M4BJS1 Q8ITG5 Q8I0E8 A0A0P6BFI2 Q8ITG4 A0A0P5IPP8 A0A2M4AK32
A0A2A4JMM7 H9J5X9 A0A3S2NFX7 H9J647 A0A194PGU6 A0A182MLJ2 A0A2H1WNI5 A0A2A4JNM0 A0A084VAF1 A0A194PT23 A0A182JAX9 A0A3S2PFN8 A0A194RLN8 A0A1I8JUI2 H9J2F6 G8GE17 A0A2C9GUN5 Q05KC0 A0A212ETT6 A0A194QZM0 A0A182Q9Y0 A0A0L7KQP2 A0A084W803 I4DPZ6 A0A1I8JVG1 A0A0P4Z3R1 A0A0P5EQ02 A0A1Y9H2Q7 D2DHG8 A0A1Y9IUV9 A0A2A4J232 A0A240PK82 A0A194PTD5 A0A240PPP9 A0A1I8JTD3 A0A0P5L0B8 A0A182F8R8 Q1WLP6 A0A2M4CMC1 A0A1Y9IV15 A0A0P5G2W6 A0A0P5T6B2 Q5TVT3 Q2ACC9 W5JDW2 E3SH50 A0A2J7QLT8 A0A2M4CH72 A0A1I8JUZ8 W5JL40 A0A194PUC3 A0A067R342 A0A0P5U9Q6 A0A240PLR9 A0A182X4T0 A0A2B4RUS4 A0A2M4BKV7 A0A1I8JSN8 A0A1I8JUE9 A0A182TA88 A0A1Y9H2R4 A0A182YM73 A0A0P5WBB6 A0A2M4BJ94 W5JFE9 A0A2J7QLX8 Q7Q503 A0A182I9E2 A0A0P5ZDU6 A0A182LQ72 A0A0N8E2B3 Q2ABY2 A0A1S3HMF6 A0A0N8D2I0 A0A1I8JVV1 Q01158 A0A1Q2TT53 H1AD94 A0A0N8ADN0 A0A182Q5M2 A0A0P4ZJZ9 A0A2M3Z9G4 A0A0T6B4U7 A0A0B7BDP1 A0A0P5EAE4 A0A1I8JVD9 Q8T6U3 A0A2M4BJS1 Q8ITG5 Q8I0E8 A0A0P6BFI2 Q8ITG4 A0A0P5IPP8 A0A2M4AK32
PDB
6AC3
E-value=8.79983e-50,
Score=492
Ontologies
GO
Topology
Subcellular location
Peroxisome
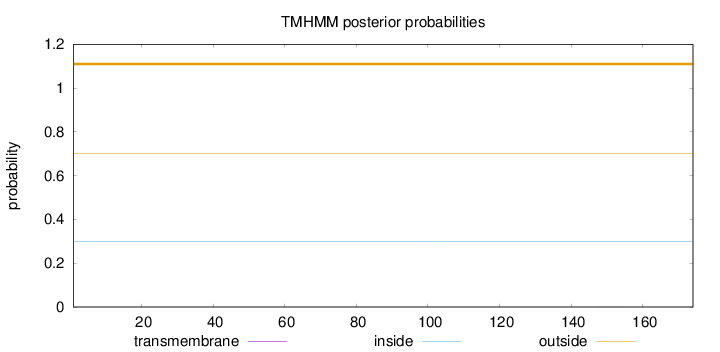
Length:
174
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00234
Exp number, first 60 AAs:
0
Total prob of N-in:
0.30016
outside
1 - 174
Population Genetic Test Statistics
Pi
210.948193
Theta
42.997402
Tajima's D
0.661629
CLR
0.544879
CSRT
0.566621668916554
Interpretation
Uncertain
