Gene
KWMTBOMO15138
Pre Gene Modal
BGIBMGA004921
Annotation
PREDICTED:_4-coumarate--CoA_ligase_1-like_[Bombyx_mori]
Full name
Luciferin 4-monooxygenase
Location in the cell
Cytoplasmic Reliability : 2.927
Sequence
CDS
ATGGTGTACTCACCTGTAGATTACTTAAAAGCCATTGAGGAATATAAGGTAAACCTTTTGATAATAGTGCCGCCAATCGCTGTTTTTCTATCAAAGTCTGATCTCGTCCAAAAGTATGACTTGTCTTCAGTGACTAGAATTTGGTGTGGTGCAGCTCCACTCAGTCAAGAGACGACTGTGGGGGTTTTAAGGAATTTCCCTAACTGTCAGAAGATAATCCATGGATACGGAATGACGGAAACATCTTTTATAATAATAAAAAGCTATGATACCTTGGACAGCGAACAAAAGCATGGAAGTTGCGGGCACGTCGTACCTGGCATGAAAGTGAAGGTTGTTGACATTAATACTCGACAAACATTAGGATCGTATGAACGAGGTGAAGTCTGCGTTAAAGGACCCCTGGTGATGAAAGGCTACATGAATAACGATCAAGCTAATGTAGATATAAGGGACGACGAGGGTTTCCTCAAAACTGGAGATATCGGATATTATGATTCAGACGGCTGTTTCTACATAGTCGACAGACTGAAAGAACTTATCAAATACAAAGGACATCAGGTAGCTCCGGCAATGATAGAGCGCGTACTGCTGCAGCACAGCGACGTGTTGGAGTGCGGAGTGGTGGGAGCGCCCGACGAGTTAGCTGGCGAGCTGCCCACCGCCTTCGTAGTGCCCAAGCCTGACGTCACGCTCACAGAAAAAGAACTGTTAGACTTTACTAATGAACGGGTCGGTAACATAGTTTGA
Protein
MVYSPVDYLKAIEEYKVNLLIIVPPIAVFLSKSDLVQKYDLSSVTRIWCGAAPLSQETTVGVLRNFPNCQKIIHGYGMTETSFIIIKSYDTLDSEQKHGSCGHVVPGMKVKVVDINTRQTLGSYERGEVCVKGPLVMKGYMNNDQANVDIRDDEGFLKTGDIGYYDSDGCFYIVDRLKELIKYKGHQVAPAMIERVLLQHSDVLECGVVGAPDELAGELPTAFVVPKPDVTLTEKELLDFTNERVGNIV
Summary
Description
Produces green light.
Produces green light with a wavelength of 544 nm.
Produces green light with a wavelength of 570 nm.
Produces green light with a wavelength of 544 nm.
Produces green light with a wavelength of 570 nm.
Catalytic Activity
ATP + firefly D-luciferin + O2 = AMP + CO2 + diphosphate + firefly oxyluciferin + hnu
Cofactor
Mg(2+)
Biophysicochemical Properties
11.5 uM for luciferin
130 uM for ATP
130 uM for ATP
Subunit
Homodimer.
Similarity
Belongs to the ATP-dependent AMP-binding enzyme family.
Keywords
ATP-binding
Luminescence
Magnesium
Metal-binding
Monooxygenase
Nucleotide-binding
Oxidoreductase
Peroxisome
Photoprotein
3D-structure
Feature
chain Luciferin 4-monooxygenase
Uniprot
H9J5Y0
H9J5X9
A0A2A4JN67
A0A2A4JMM7
A0A194PHR8
A0A194RK67
+ More
A0A3S2NFX7 A0A2H1WNI5 A0A2A4JNM0 A0A194PGU6 A0A194RLN8 A0A3S2PFN8 H9J647 A0A1Y9H2Q7 A0A2C9GUN5 A0A240PK82 A0A182MLJ2 Q2ACC9 A0A1Y9IUV9 H9J2F6 A0A1I8JUE9 A0A194PT23 G8GE17 A0A182Q5M2 I4DPZ6 A0A212ETT6 A0A240PLR9 A0A182R329 A0A182TA88 A0A182YM73 Q1WLP6 A0A1I8JUI2 A0A194QZM0 A0A182TRT7 A0A182LYC6 A0A0J7KT57 A0A182Q9Y0 A0A2J7QLT8 A0A1I8JVD9 A0A1I8JTE0 F5HJ76 A0A084VAF1 A0A182WW21 K7J1G6 A0A182JAX9 A0A1Y9H2R4 Q8ITG4 Q8T6U3 A0A1Q2TT53 Q8ITG5 Q8I0E8 A0A2J7QLX8 H1AD94 Q01158 H1AD95 E3WI50 A0A1Q2TT48 P13129 Q2ACC8 Q27348 A0A182T271 Q2ABY2 A0A084VAF2 A0A0C6DRW5 A0A2W1BTP3 A0A182YH13 A0A182W5X3 A0A1Y9IV15 A0A182YM74 Q27321 Q8IRZ9 Q25118 Q8ITG3 E2AA43 A0A232FI37 Q9U4U7 D2DHG8 E3SH50 Q26304 E0VSL5 A0A1I8JVG1 Q1AG35 B3TMS5 A0A084W803 A0A2M4CMC1 A0A182QSE0 A0A1I8JTD3 A0A2M4BJS1 A0A2M4BJM2 A0A2M4BJN0 D2DHG9 A0A240PNB0 Q5TVT3 A0A067R342 G9M6I9 A0A1I8JVV1 A0A2S3QZT6 A0A182F8R8 Q05KC0
A0A3S2NFX7 A0A2H1WNI5 A0A2A4JNM0 A0A194PGU6 A0A194RLN8 A0A3S2PFN8 H9J647 A0A1Y9H2Q7 A0A2C9GUN5 A0A240PK82 A0A182MLJ2 Q2ACC9 A0A1Y9IUV9 H9J2F6 A0A1I8JUE9 A0A194PT23 G8GE17 A0A182Q5M2 I4DPZ6 A0A212ETT6 A0A240PLR9 A0A182R329 A0A182TA88 A0A182YM73 Q1WLP6 A0A1I8JUI2 A0A194QZM0 A0A182TRT7 A0A182LYC6 A0A0J7KT57 A0A182Q9Y0 A0A2J7QLT8 A0A1I8JVD9 A0A1I8JTE0 F5HJ76 A0A084VAF1 A0A182WW21 K7J1G6 A0A182JAX9 A0A1Y9H2R4 Q8ITG4 Q8T6U3 A0A1Q2TT53 Q8ITG5 Q8I0E8 A0A2J7QLX8 H1AD94 Q01158 H1AD95 E3WI50 A0A1Q2TT48 P13129 Q2ACC8 Q27348 A0A182T271 Q2ABY2 A0A084VAF2 A0A0C6DRW5 A0A2W1BTP3 A0A182YH13 A0A182W5X3 A0A1Y9IV15 A0A182YM74 Q27321 Q8IRZ9 Q25118 Q8ITG3 E2AA43 A0A232FI37 Q9U4U7 D2DHG8 E3SH50 Q26304 E0VSL5 A0A1I8JVG1 Q1AG35 B3TMS5 A0A084W803 A0A2M4CMC1 A0A182QSE0 A0A1I8JTD3 A0A2M4BJS1 A0A2M4BJM2 A0A2M4BJN0 D2DHG9 A0A240PNB0 Q5TVT3 A0A067R342 G9M6I9 A0A1I8JVV1 A0A2S3QZT6 A0A182F8R8 Q05KC0
EC Number
1.13.12.7
Pubmed
EMBL
BABH01029976
BABH01029979
BABH01029980
NWSH01001030
PCG73034.1
PCG73036.1
+ More
KQ459604 KPI92264.1 KQ460045 KPJ18228.1 RSAL01000053 RVE50075.1 ODYU01009511 SOQ54004.1 PCG73033.1 KPI92263.1 KPJ18230.1 RVE50074.1 BABH01029982 AXCM01004086 AB196455 BAE80728.1 BABH01007891 KQ459593 KPI96566.1 JN127350 AET05796.1 AXCN02000090 AK403861 BAM19986.1 AGBW02012531 OWR44889.1 DQ137139 AAZ74651.1 KQ460890 KPJ10988.1 AXCM01007305 AXCM01007306 LBMM01003413 KMQ93562.1 NEVH01013241 PNF29543.1 APCN01002222 AAAB01008820 EGK96337.1 ATLV01003206 KE524124 KFB34945.1 AF486801 AAN40976.1 AF420006 AAM00429.1 AB909130 BAW94791.1 AF486800 AAN40975.1 AF486802 AF486803 AAN40977.1 PNF29548.1 AB644226 BAL46510.1 X66919 AB644227 BAL46511.1 AB603803 BAJ41367.1 AB908263 BAW94790.1 M26194 AB196456 BAE80729.1 U49182 AY192561 Z49891 AAA91472.1 AAO39674.1 CAA90072.1 AB220162 BAE80731.1 KFB34946.1 AB251887 BAQ25864.1 KZ149903 PZC78438.1 U49181 U51019 Z69619 AAA91471.1 AAB00229.1 CAA93444.1 AY181996 AY192560 AAN73267.1 AAO39673.2 L39929 AAC37253.1 AF486804 AAN40979.1 GL438029 EFN69681.1 NNAY01000193 OXU30119.1 AF139645 AAD34543.2 FJ545728 ACT68596.1 GU062703 ADK56478.1 S61961 DS235751 EEB16371.1 DQ138966 ABA03040.1 EU302126 ABZ88151.1 ATLV01021336 KE525316 KFB46347.1 GGFL01002163 MBW66341.1 GGFJ01004113 MBW53254.1 GGFJ01004114 MBW53255.1 GGFJ01004116 MBW53257.1 FJ545729 ACT68597.1 EAL41501.4 KK852730 KDR17523.1 AB604790 BAL40875.1 PDGH01000119 POB45175.1 AB255748 BAF34360.1
KQ459604 KPI92264.1 KQ460045 KPJ18228.1 RSAL01000053 RVE50075.1 ODYU01009511 SOQ54004.1 PCG73033.1 KPI92263.1 KPJ18230.1 RVE50074.1 BABH01029982 AXCM01004086 AB196455 BAE80728.1 BABH01007891 KQ459593 KPI96566.1 JN127350 AET05796.1 AXCN02000090 AK403861 BAM19986.1 AGBW02012531 OWR44889.1 DQ137139 AAZ74651.1 KQ460890 KPJ10988.1 AXCM01007305 AXCM01007306 LBMM01003413 KMQ93562.1 NEVH01013241 PNF29543.1 APCN01002222 AAAB01008820 EGK96337.1 ATLV01003206 KE524124 KFB34945.1 AF486801 AAN40976.1 AF420006 AAM00429.1 AB909130 BAW94791.1 AF486800 AAN40975.1 AF486802 AF486803 AAN40977.1 PNF29548.1 AB644226 BAL46510.1 X66919 AB644227 BAL46511.1 AB603803 BAJ41367.1 AB908263 BAW94790.1 M26194 AB196456 BAE80729.1 U49182 AY192561 Z49891 AAA91472.1 AAO39674.1 CAA90072.1 AB220162 BAE80731.1 KFB34946.1 AB251887 BAQ25864.1 KZ149903 PZC78438.1 U49181 U51019 Z69619 AAA91471.1 AAB00229.1 CAA93444.1 AY181996 AY192560 AAN73267.1 AAO39673.2 L39929 AAC37253.1 AF486804 AAN40979.1 GL438029 EFN69681.1 NNAY01000193 OXU30119.1 AF139645 AAD34543.2 FJ545728 ACT68596.1 GU062703 ADK56478.1 S61961 DS235751 EEB16371.1 DQ138966 ABA03040.1 EU302126 ABZ88151.1 ATLV01021336 KE525316 KFB46347.1 GGFL01002163 MBW66341.1 GGFJ01004113 MBW53254.1 GGFJ01004114 MBW53255.1 GGFJ01004116 MBW53257.1 FJ545729 ACT68597.1 EAL41501.4 KK852730 KDR17523.1 AB604790 BAL40875.1 PDGH01000119 POB45175.1 AB255748 BAF34360.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000283053
UP000075884
+ More
UP000075883 UP000075885 UP000075920 UP000075900 UP000075886 UP000007151 UP000075881 UP000075901 UP000076408 UP000075902 UP000036403 UP000235965 UP000075903 UP000075840 UP000007062 UP000030765 UP000076407 UP000002358 UP000075880 UP000000311 UP000215335 UP000009046 UP000027135 UP000237466 UP000069272
UP000075883 UP000075885 UP000075920 UP000075900 UP000075886 UP000007151 UP000075881 UP000075901 UP000076408 UP000075902 UP000036403 UP000235965 UP000075903 UP000075840 UP000007062 UP000030765 UP000076407 UP000002358 UP000075880 UP000000311 UP000215335 UP000009046 UP000027135 UP000237466 UP000069272
Interpro
Gene 3D
ProteinModelPortal
H9J5Y0
H9J5X9
A0A2A4JN67
A0A2A4JMM7
A0A194PHR8
A0A194RK67
+ More
A0A3S2NFX7 A0A2H1WNI5 A0A2A4JNM0 A0A194PGU6 A0A194RLN8 A0A3S2PFN8 H9J647 A0A1Y9H2Q7 A0A2C9GUN5 A0A240PK82 A0A182MLJ2 Q2ACC9 A0A1Y9IUV9 H9J2F6 A0A1I8JUE9 A0A194PT23 G8GE17 A0A182Q5M2 I4DPZ6 A0A212ETT6 A0A240PLR9 A0A182R329 A0A182TA88 A0A182YM73 Q1WLP6 A0A1I8JUI2 A0A194QZM0 A0A182TRT7 A0A182LYC6 A0A0J7KT57 A0A182Q9Y0 A0A2J7QLT8 A0A1I8JVD9 A0A1I8JTE0 F5HJ76 A0A084VAF1 A0A182WW21 K7J1G6 A0A182JAX9 A0A1Y9H2R4 Q8ITG4 Q8T6U3 A0A1Q2TT53 Q8ITG5 Q8I0E8 A0A2J7QLX8 H1AD94 Q01158 H1AD95 E3WI50 A0A1Q2TT48 P13129 Q2ACC8 Q27348 A0A182T271 Q2ABY2 A0A084VAF2 A0A0C6DRW5 A0A2W1BTP3 A0A182YH13 A0A182W5X3 A0A1Y9IV15 A0A182YM74 Q27321 Q8IRZ9 Q25118 Q8ITG3 E2AA43 A0A232FI37 Q9U4U7 D2DHG8 E3SH50 Q26304 E0VSL5 A0A1I8JVG1 Q1AG35 B3TMS5 A0A084W803 A0A2M4CMC1 A0A182QSE0 A0A1I8JTD3 A0A2M4BJS1 A0A2M4BJM2 A0A2M4BJN0 D2DHG9 A0A240PNB0 Q5TVT3 A0A067R342 G9M6I9 A0A1I8JVV1 A0A2S3QZT6 A0A182F8R8 Q05KC0
A0A3S2NFX7 A0A2H1WNI5 A0A2A4JNM0 A0A194PGU6 A0A194RLN8 A0A3S2PFN8 H9J647 A0A1Y9H2Q7 A0A2C9GUN5 A0A240PK82 A0A182MLJ2 Q2ACC9 A0A1Y9IUV9 H9J2F6 A0A1I8JUE9 A0A194PT23 G8GE17 A0A182Q5M2 I4DPZ6 A0A212ETT6 A0A240PLR9 A0A182R329 A0A182TA88 A0A182YM73 Q1WLP6 A0A1I8JUI2 A0A194QZM0 A0A182TRT7 A0A182LYC6 A0A0J7KT57 A0A182Q9Y0 A0A2J7QLT8 A0A1I8JVD9 A0A1I8JTE0 F5HJ76 A0A084VAF1 A0A182WW21 K7J1G6 A0A182JAX9 A0A1Y9H2R4 Q8ITG4 Q8T6U3 A0A1Q2TT53 Q8ITG5 Q8I0E8 A0A2J7QLX8 H1AD94 Q01158 H1AD95 E3WI50 A0A1Q2TT48 P13129 Q2ACC8 Q27348 A0A182T271 Q2ABY2 A0A084VAF2 A0A0C6DRW5 A0A2W1BTP3 A0A182YH13 A0A182W5X3 A0A1Y9IV15 A0A182YM74 Q27321 Q8IRZ9 Q25118 Q8ITG3 E2AA43 A0A232FI37 Q9U4U7 D2DHG8 E3SH50 Q26304 E0VSL5 A0A1I8JVG1 Q1AG35 B3TMS5 A0A084W803 A0A2M4CMC1 A0A182QSE0 A0A1I8JTD3 A0A2M4BJS1 A0A2M4BJM2 A0A2M4BJN0 D2DHG9 A0A240PNB0 Q5TVT3 A0A067R342 G9M6I9 A0A1I8JVV1 A0A2S3QZT6 A0A182F8R8 Q05KC0
PDB
6AC3
E-value=1.55295e-62,
Score=605
Ontologies
GO
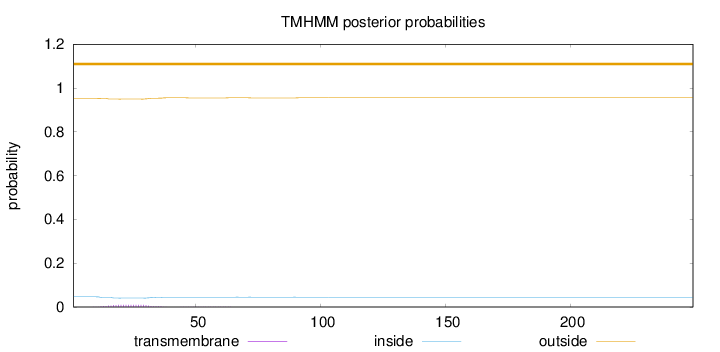
Topology
Subcellular location
Peroxisome
Length:
249
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.18782
Exp number, first 60 AAs:
0.18544
Total prob of N-in:
0.04645
outside
1 - 249
Population Genetic Test Statistics
Pi
155.482437
Theta
163.166763
Tajima's D
0.377845
CLR
0.269612
CSRT
0.476626168691565
Interpretation
Uncertain
