Pre Gene Modal
BGIBMGA004920
Annotation
PREDICTED:_luciferin_4-monooxygenase-like_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.594
Sequence
CDS
ATGAAAAAAATTGGCGTGAAGCGTGGAGATGTCATAGCATTCTGTGGCGAAAATATAAATGAATTCTTTATAGGTATCTTGAGCACTATTTGCTGTGGTGCCACTGTTACAACATTGAATGTAATGTACTCGGAAGATGAAATCAATCACGTTTTGAATATATCAAAACCAAGTATGATATTTGGTTCTGAAATAGCATTCAAAAGAAATTTGGGTGTTTTCAAATCACTACCCTTCATCAAAAATTTTGTACAATTGAACGGCAGTCCAGTGGAGCGGGGAATCCAGACCTTCAGCTCGTTATTGCTGCCAACCGATCCTAATATGTACGAGCCGGCACTAGAGGCCCGTGGCGATGACACAGCATTCATTCTATATTCTTCAGGCACAACTGGTCTGCCTAAAGGAGTAATGCTAACACATCTCAACGCTTTGTATACTTCAGAAGTTTTTGAGCACGGTGAATTGAATGTTGTTGATAAAAGTTGTTACACTTTGCTTACCATCGTGCCATGGTATCATGCGTATGGATTGATGTCAACTATAAATAATGTACTTTTAAAGAATACTTCGGTTTATTTGTCTGGTTTTAATCCTTTGCAGTACTTGAAGTGTATTCAAGATTATAAGGTCAATGTTTTATTAGTTGTTCCGCCTATAGTCGTGTTTTTAGCGAAGGCTCCAATTGTAGGAAAATTTGATTTATCTTCAGTTAACAAAATATGGTGTGGTGCCGCTCCTCTTAATTCGGAAACTATTAATGAAACTCTGAAAAGACTGCCAAACTGTGAAGGAATTTTCCAAGGATACGGTATGACGGAGACTTCTGTGGCGGCGACAAAAGATATAAACAAAGATGGTGTTGTGCGTAAGCCTGGCAGTGGAGGTTATGTTCTGCCGGGCCTTCGTGCCAAGGTGGTAAATATAGAAACCCGTGAAAGATTGGGACCGTATTGCGAAGGTGAAATTTGCTTCAAAGGACCCGTAATAATGAAAGGTTATGCAAATAACAAAAAAGCAACTGCTGAAATAACTGACGATGAAGGGTATCTAAATACTGGAGACATCGGATACTACGATGATGACGGCTGCTTCTTCGTTGTGGATCGACTGAAAGAACTCATCAAATACAAAGGACACCAGGTAGCCCCAGCAATAATAGAGCGCGTACTGCTGCAACACAGCGACGTGTTGGAGTGCGGAGTGGTGGGAGCGCCCGACGAGTTGGCTGGCGAGCTGCCCACCGCCTTCGTAGTGCCCAAGCCTGACGTCACGCTCACAGAAAGAGAACTGCTAGACTTTACTAATGAACGGTTGTCATCTACATCAAGACTCCACGGTGGTATTATATTCGTCAAAGAAATACCAAAAAACGCGTCGGGAAAAATACTGCGGCGTGTACTCAAACAAAGAATTGCAGAAATGAAGAAAAGCAAATTATAA
Protein
MKKIGVKRGDVIAFCGENINEFFIGILSTICCGATVTTLNVMYSEDEINHVLNISKPSMIFGSEIAFKRNLGVFKSLPFIKNFVQLNGSPVERGIQTFSSLLLPTDPNMYEPALEARGDDTAFILYSSGTTGLPKGVMLTHLNALYTSEVFEHGELNVVDKSCYTLLTIVPWYHAYGLMSTINNVLLKNTSVYLSGFNPLQYLKCIQDYKVNVLLVVPPIVVFLAKAPIVGKFDLSSVNKIWCGAAPLNSETINETLKRLPNCEGIFQGYGMTETSVAATKDINKDGVVRKPGSGGYVLPGLRAKVVNIETRERLGPYCEGEICFKGPVIMKGYANNKKATAEITDDEGYLNTGDIGYYDDDGCFFVVDRLKELIKYKGHQVAPAIIERVLLQHSDVLECGVVGAPDELAGELPTAFVVPKPDVTLTERELLDFTNERLSSTSRLHGGIIFVKEIPKNASGKILRRVLKQRIAEMKKSKL
Summary
Uniprot
H9J5X9
A0A2A4JMM7
A0A2A4JN67
A0A3S2NFX7
A0A194PHR8
A0A194RK67
+ More
A0A3S2PFN8 A0A194QZM0 A0A194PT23 A0A212ETT6 A0A2A4JNM0 A0A194PGU6 A0A212FII9 A0A2H1WNI5 A0A194RLN8 A0A0L0CRE8 A0A1L8EBZ5 A0A1L8ECA8 B3M0L3 A0A1I8N0L6 A0A1L8EBY4 A0A3B0JK27 T1P9V4 B3P785 I4DPZ6 A0A084W803 A0A0C6E621 A0A194QZV5 B4MBY1 A0A182Q9Y0 Q9VCC6 B4G408 A0A0A1X815 Q298J5 A0A2H1VTP5 A0A182MLJ2 H9J2F6 A0A1Y9H2R4 G8GE17 B4K7E6 A0A2A4JYH7 A0A1L8DYF7 K7J1G6 B4NKS0 A0A1Y9IV15 A0A2W1BTP3 A0A1L8DYB2 A0A182QSE0 W5JL40 B4PL10 A0A194PZH5 B4QSH4 A0A212FIH9 B4JIF6 A0A182JAX9 A0A182W5X3 A0A1I8JUI2 A0A1I8JVG1 A0A1A9VQZ3 A0A1I8PLT1 A0A1I8PLP9 A0A1W4V678 W8AT94 A0A2M4AKM3 A0A1B0DFB0 A0A182F8R8 U5EXU9 B4JIF8 A0A182T271 Q05KC0 A0A2M4AK32 A0A1L8DYZ5 A0A1L8DZF9 A0A2A4J232 Q5TVT3 A0A1I8JTD3 A0A182PST3 A0A182R329 A0A2M4BKV7 D3TNT8 A0A0M5JC90 A0A1Q3FIY4 A0A0C6DRW5 A0A1A9ZLB4 A0A2M4BJ94 A0A182QGS8 A0A182SUG3 H9JJH4 A0A034WF13 B0W9Q1 A0A240PPP9 A0A1A9XI15 F4WWH1 A0A1I8JVV1 A0A084VFI3 A9ZPM4 A0A1B0DRC8 A0A1Y9IVT1 A0A1I8JUZ8
A0A3S2PFN8 A0A194QZM0 A0A194PT23 A0A212ETT6 A0A2A4JNM0 A0A194PGU6 A0A212FII9 A0A2H1WNI5 A0A194RLN8 A0A0L0CRE8 A0A1L8EBZ5 A0A1L8ECA8 B3M0L3 A0A1I8N0L6 A0A1L8EBY4 A0A3B0JK27 T1P9V4 B3P785 I4DPZ6 A0A084W803 A0A0C6E621 A0A194QZV5 B4MBY1 A0A182Q9Y0 Q9VCC6 B4G408 A0A0A1X815 Q298J5 A0A2H1VTP5 A0A182MLJ2 H9J2F6 A0A1Y9H2R4 G8GE17 B4K7E6 A0A2A4JYH7 A0A1L8DYF7 K7J1G6 B4NKS0 A0A1Y9IV15 A0A2W1BTP3 A0A1L8DYB2 A0A182QSE0 W5JL40 B4PL10 A0A194PZH5 B4QSH4 A0A212FIH9 B4JIF6 A0A182JAX9 A0A182W5X3 A0A1I8JUI2 A0A1I8JVG1 A0A1A9VQZ3 A0A1I8PLT1 A0A1I8PLP9 A0A1W4V678 W8AT94 A0A2M4AKM3 A0A1B0DFB0 A0A182F8R8 U5EXU9 B4JIF8 A0A182T271 Q05KC0 A0A2M4AK32 A0A1L8DYZ5 A0A1L8DZF9 A0A2A4J232 Q5TVT3 A0A1I8JTD3 A0A182PST3 A0A182R329 A0A2M4BKV7 D3TNT8 A0A0M5JC90 A0A1Q3FIY4 A0A0C6DRW5 A0A1A9ZLB4 A0A2M4BJ94 A0A182QGS8 A0A182SUG3 H9JJH4 A0A034WF13 B0W9Q1 A0A240PPP9 A0A1A9XI15 F4WWH1 A0A1I8JVV1 A0A084VFI3 A9ZPM4 A0A1B0DRC8 A0A1Y9IVT1 A0A1I8JUZ8
Pubmed
19121390
26354079
22118469
26108605
17994087
18057021
+ More
25315136 22651552 24438588 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25830018 15632085 23185243 20075255 28756777 20920257 23761445 17550304 24495485 12364791 14747013 17210077 20353571 25348373 21719571 17996401
25315136 22651552 24438588 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25830018 15632085 23185243 20075255 28756777 20920257 23761445 17550304 24495485 12364791 14747013 17210077 20353571 25348373 21719571 17996401
EMBL
BABH01029979
BABH01029980
NWSH01001030
PCG73036.1
PCG73034.1
RSAL01000053
+ More
RVE50075.1 KQ459604 KPI92264.1 KQ460045 KPJ18228.1 RVE50074.1 KQ460890 KPJ10988.1 KQ459593 KPI96566.1 AGBW02012531 OWR44889.1 PCG73033.1 KPI92263.1 AGBW02008380 OWR53544.1 ODYU01009511 SOQ54004.1 KPJ18230.1 JRES01000028 KNC34816.1 GFDG01002539 JAV16260.1 GFDG01002537 JAV16262.1 CH902617 EDV44260.1 KPU80729.1 GFDG01002538 JAV16261.1 OUUW01000007 SPP82704.1 KA645541 AFP60170.1 CH954182 EDV53905.1 AK403861 BAM19986.1 ATLV01021336 KE525316 KFB46347.1 AB251886 BAQ25863.1 KPJ10987.1 CH940656 EDW58602.1 KRF78360.1 AXCN02000090 AE014297 AY060906 AY121681 AAF56245.1 AAL28454.1 AAM52008.1 CH479179 EDW24419.1 GBXI01007236 JAD07056.1 CM000070 EAL27960.1 KRT00363.1 ODYU01004352 SOQ44136.1 AXCM01004086 BABH01007891 JN127350 AET05796.1 CH933806 EDW14270.1 NWSH01000380 PCG76826.1 GFDF01002635 JAV11449.1 CH964272 EDW84131.1 KZ149903 PZC78438.1 GFDF01002634 JAV11450.1 ADMH02001214 ETN63630.1 CM000160 EDW98995.1 KPI96565.1 CM000364 EDX14173.1 OWR53543.1 CH916369 EDV93037.1 GAMC01014530 GAMC01014529 JAB92025.1 GGFK01007847 MBW41168.1 AJVK01015100 AJVK01015101 AJVK01015102 AJVK01015103 GANO01000766 JAB59105.1 EDV93039.1 AB255748 BAF34360.1 GGFK01007825 MBW41146.1 GFDF01002420 JAV11664.1 GFDF01002419 JAV11665.1 NWSH01003686 PCG65919.1 AAAB01008820 EAL41501.4 APCN01002222 GGFJ01004568 MBW53709.1 CCAG010000200 EZ423090 ADD19366.1 CP012526 ALC45981.1 GFDL01007475 JAV27570.1 AB251887 BAQ25864.1 GGFJ01003984 MBW53125.1 AXCN02001407 BABH01017566 BABH01017567 GAKP01004796 JAC54156.1 DS231865 EDS40370.1 GL888406 EGI61513.1 ATLV01012411 KE524791 KFB36727.1 AB353886 BAF96580.1 AJVK01019897 AJVK01019898 AJVK01019899 AJVK01019900
RVE50075.1 KQ459604 KPI92264.1 KQ460045 KPJ18228.1 RVE50074.1 KQ460890 KPJ10988.1 KQ459593 KPI96566.1 AGBW02012531 OWR44889.1 PCG73033.1 KPI92263.1 AGBW02008380 OWR53544.1 ODYU01009511 SOQ54004.1 KPJ18230.1 JRES01000028 KNC34816.1 GFDG01002539 JAV16260.1 GFDG01002537 JAV16262.1 CH902617 EDV44260.1 KPU80729.1 GFDG01002538 JAV16261.1 OUUW01000007 SPP82704.1 KA645541 AFP60170.1 CH954182 EDV53905.1 AK403861 BAM19986.1 ATLV01021336 KE525316 KFB46347.1 AB251886 BAQ25863.1 KPJ10987.1 CH940656 EDW58602.1 KRF78360.1 AXCN02000090 AE014297 AY060906 AY121681 AAF56245.1 AAL28454.1 AAM52008.1 CH479179 EDW24419.1 GBXI01007236 JAD07056.1 CM000070 EAL27960.1 KRT00363.1 ODYU01004352 SOQ44136.1 AXCM01004086 BABH01007891 JN127350 AET05796.1 CH933806 EDW14270.1 NWSH01000380 PCG76826.1 GFDF01002635 JAV11449.1 CH964272 EDW84131.1 KZ149903 PZC78438.1 GFDF01002634 JAV11450.1 ADMH02001214 ETN63630.1 CM000160 EDW98995.1 KPI96565.1 CM000364 EDX14173.1 OWR53543.1 CH916369 EDV93037.1 GAMC01014530 GAMC01014529 JAB92025.1 GGFK01007847 MBW41168.1 AJVK01015100 AJVK01015101 AJVK01015102 AJVK01015103 GANO01000766 JAB59105.1 EDV93039.1 AB255748 BAF34360.1 GGFK01007825 MBW41146.1 GFDF01002420 JAV11664.1 GFDF01002419 JAV11665.1 NWSH01003686 PCG65919.1 AAAB01008820 EAL41501.4 APCN01002222 GGFJ01004568 MBW53709.1 CCAG010000200 EZ423090 ADD19366.1 CP012526 ALC45981.1 GFDL01007475 JAV27570.1 AB251887 BAQ25864.1 GGFJ01003984 MBW53125.1 AXCN02001407 BABH01017566 BABH01017567 GAKP01004796 JAC54156.1 DS231865 EDS40370.1 GL888406 EGI61513.1 ATLV01012411 KE524791 KFB36727.1 AB353886 BAF96580.1 AJVK01019897 AJVK01019898 AJVK01019899 AJVK01019900
Proteomes
UP000005204
UP000218220
UP000283053
UP000053268
UP000053240
UP000007151
+ More
UP000037069 UP000007801 UP000095301 UP000268350 UP000008711 UP000030765 UP000008792 UP000075886 UP000000803 UP000008744 UP000001819 UP000075883 UP000075884 UP000009192 UP000002358 UP000007798 UP000075920 UP000000673 UP000002282 UP000000304 UP000001070 UP000075880 UP000075900 UP000075903 UP000078200 UP000095300 UP000192221 UP000092462 UP000069272 UP000075901 UP000007062 UP000075840 UP000075885 UP000092444 UP000092553 UP000092445 UP000002320 UP000092443 UP000007755 UP000076407 UP000075902
UP000037069 UP000007801 UP000095301 UP000268350 UP000008711 UP000030765 UP000008792 UP000075886 UP000000803 UP000008744 UP000001819 UP000075883 UP000075884 UP000009192 UP000002358 UP000007798 UP000075920 UP000000673 UP000002282 UP000000304 UP000001070 UP000075880 UP000075900 UP000075903 UP000078200 UP000095300 UP000192221 UP000092462 UP000069272 UP000075901 UP000007062 UP000075840 UP000075885 UP000092444 UP000092553 UP000092445 UP000002320 UP000092443 UP000007755 UP000076407 UP000075902
Interpro
Gene 3D
ProteinModelPortal
H9J5X9
A0A2A4JMM7
A0A2A4JN67
A0A3S2NFX7
A0A194PHR8
A0A194RK67
+ More
A0A3S2PFN8 A0A194QZM0 A0A194PT23 A0A212ETT6 A0A2A4JNM0 A0A194PGU6 A0A212FII9 A0A2H1WNI5 A0A194RLN8 A0A0L0CRE8 A0A1L8EBZ5 A0A1L8ECA8 B3M0L3 A0A1I8N0L6 A0A1L8EBY4 A0A3B0JK27 T1P9V4 B3P785 I4DPZ6 A0A084W803 A0A0C6E621 A0A194QZV5 B4MBY1 A0A182Q9Y0 Q9VCC6 B4G408 A0A0A1X815 Q298J5 A0A2H1VTP5 A0A182MLJ2 H9J2F6 A0A1Y9H2R4 G8GE17 B4K7E6 A0A2A4JYH7 A0A1L8DYF7 K7J1G6 B4NKS0 A0A1Y9IV15 A0A2W1BTP3 A0A1L8DYB2 A0A182QSE0 W5JL40 B4PL10 A0A194PZH5 B4QSH4 A0A212FIH9 B4JIF6 A0A182JAX9 A0A182W5X3 A0A1I8JUI2 A0A1I8JVG1 A0A1A9VQZ3 A0A1I8PLT1 A0A1I8PLP9 A0A1W4V678 W8AT94 A0A2M4AKM3 A0A1B0DFB0 A0A182F8R8 U5EXU9 B4JIF8 A0A182T271 Q05KC0 A0A2M4AK32 A0A1L8DYZ5 A0A1L8DZF9 A0A2A4J232 Q5TVT3 A0A1I8JTD3 A0A182PST3 A0A182R329 A0A2M4BKV7 D3TNT8 A0A0M5JC90 A0A1Q3FIY4 A0A0C6DRW5 A0A1A9ZLB4 A0A2M4BJ94 A0A182QGS8 A0A182SUG3 H9JJH4 A0A034WF13 B0W9Q1 A0A240PPP9 A0A1A9XI15 F4WWH1 A0A1I8JVV1 A0A084VFI3 A9ZPM4 A0A1B0DRC8 A0A1Y9IVT1 A0A1I8JUZ8
A0A3S2PFN8 A0A194QZM0 A0A194PT23 A0A212ETT6 A0A2A4JNM0 A0A194PGU6 A0A212FII9 A0A2H1WNI5 A0A194RLN8 A0A0L0CRE8 A0A1L8EBZ5 A0A1L8ECA8 B3M0L3 A0A1I8N0L6 A0A1L8EBY4 A0A3B0JK27 T1P9V4 B3P785 I4DPZ6 A0A084W803 A0A0C6E621 A0A194QZV5 B4MBY1 A0A182Q9Y0 Q9VCC6 B4G408 A0A0A1X815 Q298J5 A0A2H1VTP5 A0A182MLJ2 H9J2F6 A0A1Y9H2R4 G8GE17 B4K7E6 A0A2A4JYH7 A0A1L8DYF7 K7J1G6 B4NKS0 A0A1Y9IV15 A0A2W1BTP3 A0A1L8DYB2 A0A182QSE0 W5JL40 B4PL10 A0A194PZH5 B4QSH4 A0A212FIH9 B4JIF6 A0A182JAX9 A0A182W5X3 A0A1I8JUI2 A0A1I8JVG1 A0A1A9VQZ3 A0A1I8PLT1 A0A1I8PLP9 A0A1W4V678 W8AT94 A0A2M4AKM3 A0A1B0DFB0 A0A182F8R8 U5EXU9 B4JIF8 A0A182T271 Q05KC0 A0A2M4AK32 A0A1L8DYZ5 A0A1L8DZF9 A0A2A4J232 Q5TVT3 A0A1I8JTD3 A0A182PST3 A0A182R329 A0A2M4BKV7 D3TNT8 A0A0M5JC90 A0A1Q3FIY4 A0A0C6DRW5 A0A1A9ZLB4 A0A2M4BJ94 A0A182QGS8 A0A182SUG3 H9JJH4 A0A034WF13 B0W9Q1 A0A240PPP9 A0A1A9XI15 F4WWH1 A0A1I8JVV1 A0A084VFI3 A9ZPM4 A0A1B0DRC8 A0A1Y9IVT1 A0A1I8JUZ8
PDB
2D1T
E-value=2.7637e-95,
Score=891
Ontologies
GO
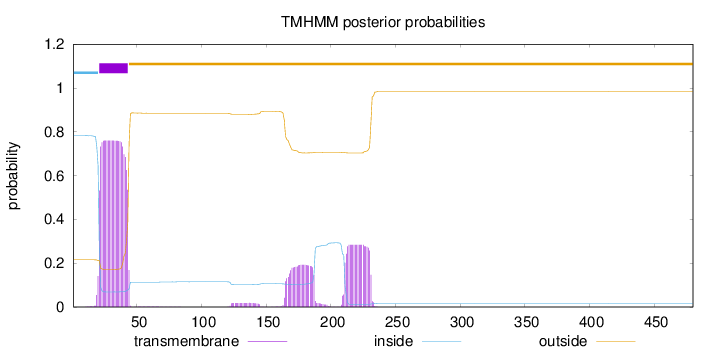
Topology
Length:
480
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
27.59187
Exp number, first 60 AAs:
17.08653
Total prob of N-in:
0.78418
POSSIBLE N-term signal
sequence
inside
1 - 20
TMhelix
21 - 43
outside
44 - 480
Population Genetic Test Statistics
Pi
213.296026
Theta
187.491149
Tajima's D
0.862247
CLR
0.016215
CSRT
0.622068896555172
Interpretation
Uncertain
