Gene
KWMTBOMO15135 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA004919
Annotation
PREDICTED:_3'(2')?5'-bisphosphate_nucleotidase_1_isoform_X2_[Papilio_xuthus]
Location in the cell
Cytoplasmic Reliability : 2.762
Sequence
CDS
ATGTACGGTAGTGTTCCGTTAATTGTTAGGTTACTAGCTTCGTCAGTATCAGTTGCCAACCGAGCTGGGAAAATCGTTCGCGATGTAATGAGCAAAGGGGAACTTGGTATAGTGGAAAAGGGTAAAGACGACTATCAAACAGAAGCAGATAGGTCTGCTCAACGCTGTATTGTTGCATCACTTGCAGCTCAATATCCTAACTTAAAAATAATAGGTGAAGAAGATAGTTTGGAGGATGAGGGTGAAGTTGTGAGTGATTGGTTAGTGAATGAAATAGATAAGGAAATATTAAAATTACAATGTCCTCCAAATCTTCAAGAAGTCAAGGAAGAGGATATTGTAGTGTGGGTGGATCCACTTGATGGAACTTCAGAATATACTCAAGGATTCTTAGAACATGTGACTGTACTAATTGGTATAGCTGTGAATGAGACACCAGTTGCTGGTGTCATCCATCAACCATACTATAAAAATATAGTTGAAGGTGACAAGAAAATCGGTAGGACAATTTGGGGACTGCATGGTGTGGGTGTAGGTGGATTTACCCCGGCTCCTCCTCCTGAATCCATCGTAATTACAACTACTAGAAGCCATTCGAATCCTGTTGTGGAGGAGGCACTAAAAGTTATGAATGCTTCAAAGGTGCTTAGGGTTGGTGGAGCTGGTTACAAGGTCCTGCAATTATTAGAAGGAAATGCATCAATATATGTTTTTGCTAGTCCCGGCTGTAAGAAATGGGATACTTGCGCACCTGAAGCGATTCTTTCCGCAGCAGGAGGGAAATTGACTGATATACTTGGTAACTTTTACAAATATGGAGCAAATGTTACTAAACCAAATAAAACTGGTGTGTTAGCTGCAGTTAATAACGAACTACACAGTTACGCTTTAGAAAGAATACCTGAAAAACTACAAGAAACTTTAGCAAATAAGTAG
Protein
MYGSVPLIVRLLASSVSVANRAGKIVRDVMSKGELGIVEKGKDDYQTEADRSAQRCIVASLAAQYPNLKIIGEEDSLEDEGEVVSDWLVNEIDKEILKLQCPPNLQEVKEEDIVVWVDPLDGTSEYTQGFLEHVTVLIGIAVNETPVAGVIHQPYYKNIVEGDKKIGRTIWGLHGVGVGGFTPAPPPESIVITTTRSHSNPVVEEALKVMNASKVLRVGGAGYKVLQLLEGNASIYVFASPGCKKWDTCAPEAILSAAGGKLTDILGNFYKYGANVTKPNKTGVLAAVNNELHSYALERIPEKLQETLANK
Summary
Uniprot
H9J5X8
A0A194PG39
A0A2H1WLP3
A0A212ETS0
A0A0L7LEW9
A0A2A4JLR9
+ More
A0A194RQC8 A0A0L0BYC9 D6WQP1 A0A084W804 A0A182YEP8 A0A1S4F9I5 A0A2P8YXD2 A0A1B0CJ88 A0A182IXC6 A0A182F8R9 A0A2M4ALZ1 A0A1I8MRT0 A0A182VPX4 A0A2M4BV34 A0A1L8ECR1 W5JHP7 A0A182QXQ2 Q17B69 A0A182VB10 A0A182XRT0 Q7Q502 A0A182RE73 E0VSL4 A0A1L8DMG7 U5EYU6 A0A182NN51 A0A182LQ68 A0A1B0CYY1 A0A067QMS0 A0A1Q3FGU0 Q9VAG9 A0A154P4T9 A0A182JWZ2 A0A1Y1MF75 A0A1I8P0S4 B4KAN4 B0W9Q0 B4M0P9 B3LWX7 B4HZF3 B3P5C8 B4PPJ7 B4JRJ7 D3TMF5 Q297T3 A0A1W4XCE3 A0A3B0JJT5 A0A0K8TP35 A0A1B0B300 K7J1G5 B4G305 A0A232FHF8 A0A240SWM1 A0A1B6E0N3 A0A1B6E0G8 A0A195FRX8 A0A0M3QY31 E2BCN0 A0A1W4UR76 F4WDD9 A0A3L8DMB2 T1DQG2 A0A026W8F8 A0A151IX35 A0A1A9VDQ5 A0A0N8CT24 A0A0L7R1S5 A0A182G572 A0A2A3ERP2 A0A1A9W5P8 A0A151XDH1 E9IZR7 A0A195CBZ0 A0A2J7R6G7 A0A0M8ZSQ1 R7U4I7 E9FWX6 E2AVH5 B4NBM3 A0A158NIS7 A0A0P5BE40 A0A0P4YHI5 A0A0K8S789 A0A195BX77 A0A1A9ZSU8 A0A0A9XDR7 A0A0P5E012 A0A0N8CLE9 A0A1B0FLB9 W8C1J4 Q4VRW1 A0A131XXM0 A0A0P5UZF2 A0A0K8RNI9
A0A194RQC8 A0A0L0BYC9 D6WQP1 A0A084W804 A0A182YEP8 A0A1S4F9I5 A0A2P8YXD2 A0A1B0CJ88 A0A182IXC6 A0A182F8R9 A0A2M4ALZ1 A0A1I8MRT0 A0A182VPX4 A0A2M4BV34 A0A1L8ECR1 W5JHP7 A0A182QXQ2 Q17B69 A0A182VB10 A0A182XRT0 Q7Q502 A0A182RE73 E0VSL4 A0A1L8DMG7 U5EYU6 A0A182NN51 A0A182LQ68 A0A1B0CYY1 A0A067QMS0 A0A1Q3FGU0 Q9VAG9 A0A154P4T9 A0A182JWZ2 A0A1Y1MF75 A0A1I8P0S4 B4KAN4 B0W9Q0 B4M0P9 B3LWX7 B4HZF3 B3P5C8 B4PPJ7 B4JRJ7 D3TMF5 Q297T3 A0A1W4XCE3 A0A3B0JJT5 A0A0K8TP35 A0A1B0B300 K7J1G5 B4G305 A0A232FHF8 A0A240SWM1 A0A1B6E0N3 A0A1B6E0G8 A0A195FRX8 A0A0M3QY31 E2BCN0 A0A1W4UR76 F4WDD9 A0A3L8DMB2 T1DQG2 A0A026W8F8 A0A151IX35 A0A1A9VDQ5 A0A0N8CT24 A0A0L7R1S5 A0A182G572 A0A2A3ERP2 A0A1A9W5P8 A0A151XDH1 E9IZR7 A0A195CBZ0 A0A2J7R6G7 A0A0M8ZSQ1 R7U4I7 E9FWX6 E2AVH5 B4NBM3 A0A158NIS7 A0A0P5BE40 A0A0P4YHI5 A0A0K8S789 A0A195BX77 A0A1A9ZSU8 A0A0A9XDR7 A0A0P5E012 A0A0N8CLE9 A0A1B0FLB9 W8C1J4 Q4VRW1 A0A131XXM0 A0A0P5UZF2 A0A0K8RNI9
Pubmed
19121390
26354079
22118469
26227816
26108605
18362917
+ More
19820115 24438588 25244985 29403074 25315136 20920257 23761445 17510324 12364791 14747013 17210077 20566863 20966253 24845553 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 28004739 17994087 17550304 20353571 15632085 26369729 20075255 28648823 20798317 21719571 30249741 24508170 26483478 21282665 23254933 21292972 21347285 26823975 25401762 24495485 12615446 15824899
19820115 24438588 25244985 29403074 25315136 20920257 23761445 17510324 12364791 14747013 17210077 20566863 20966253 24845553 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 28004739 17994087 17550304 20353571 15632085 26369729 20075255 28648823 20798317 21719571 30249741 24508170 26483478 21282665 23254933 21292972 21347285 26823975 25401762 24495485 12615446 15824899
EMBL
BABH01029982
KQ459604
KPI92262.1
ODYU01009511
SOQ54003.1
AGBW02012531
+ More
OWR44890.1 JTDY01001414 KOB73924.1 NWSH01001030 PCG73025.1 KQ460045 KPJ18231.1 JRES01001160 KNC24976.1 KQ971354 EFA06048.1 ATLV01021336 KE525316 KFB46348.1 PYGN01000302 PSN48907.1 AJWK01014309 GGFK01008495 MBW41816.1 GGFJ01007711 MBW56852.1 GFDG01002446 JAV16353.1 ADMH02001214 ETN63631.1 AXCN02002214 CH477325 EAT43498.1 AAAB01008961 EAA11996.1 DS235751 EEB16370.1 GFDF01006519 JAV07565.1 GANO01001805 JAB58066.1 AJVK01020482 KK853153 KDR10578.1 GFDL01008255 JAV26790.1 AE014297 AY118554 AAF56941.1 AAM49923.1 KQ434809 KZC06218.1 GEZM01033253 GEZM01033252 GEZM01033251 JAV84303.1 CH933806 EDW16771.1 DS231865 EDS40369.1 CH940650 EDW67341.1 CH902617 EDV43816.1 CH480819 EDW53410.1 CH954182 EDV53178.1 CM000160 EDW98247.1 CH916373 EDV94387.1 EZ422607 ADD18883.1 CM000070 EAL28122.2 OUUW01000005 SPP81053.1 GDAI01001943 JAI15660.1 JXJN01007738 CH479179 EDW24200.1 NNAY01000193 OXU30112.1 GEDC01005861 JAS31437.1 GEDC01005866 JAS31432.1 KQ981285 KYN43203.1 CP012526 ALC46919.1 GL447336 EFN86540.1 GL888087 EGI67719.1 QOIP01000007 RLU20958.1 GAMD01002514 JAA99076.1 KK107364 EZA51931.1 KQ980832 KYN12396.1 GDIP01101655 JAM02060.1 KQ414667 KOC64741.1 JXUM01042943 KQ561344 KXJ78885.1 KZ288194 PBC33942.1 KQ982289 KYQ58424.1 GL767232 EFZ13990.1 KQ978068 KYM97608.1 NEVH01006756 PNF36424.1 KQ435863 KOX70355.1 AMQN01010616 KB308243 ELT98080.1 GL732526 EFX87971.1 GL443112 EFN62570.1 CH964232 EDW81187.1 ADTU01017112 GDIP01186102 JAJ37300.1 GDIP01230888 LRGB01001361 JAI92513.1 KZS12361.1 GBRD01016656 GDHC01008634 JAG49171.1 JAQ09995.1 KQ976394 KYM93244.1 GBHO01028369 JAG15235.1 GDIP01148506 JAJ74896.1 GDIP01120255 JAL83459.1 CCAG010013281 GAMC01003537 GAMC01003535 JAC03019.1 AY652655 AAV67032.1 GEFM01003792 JAP72004.1 GDIP01107208 JAL96506.1 GADI01001197 JAA72611.1
OWR44890.1 JTDY01001414 KOB73924.1 NWSH01001030 PCG73025.1 KQ460045 KPJ18231.1 JRES01001160 KNC24976.1 KQ971354 EFA06048.1 ATLV01021336 KE525316 KFB46348.1 PYGN01000302 PSN48907.1 AJWK01014309 GGFK01008495 MBW41816.1 GGFJ01007711 MBW56852.1 GFDG01002446 JAV16353.1 ADMH02001214 ETN63631.1 AXCN02002214 CH477325 EAT43498.1 AAAB01008961 EAA11996.1 DS235751 EEB16370.1 GFDF01006519 JAV07565.1 GANO01001805 JAB58066.1 AJVK01020482 KK853153 KDR10578.1 GFDL01008255 JAV26790.1 AE014297 AY118554 AAF56941.1 AAM49923.1 KQ434809 KZC06218.1 GEZM01033253 GEZM01033252 GEZM01033251 JAV84303.1 CH933806 EDW16771.1 DS231865 EDS40369.1 CH940650 EDW67341.1 CH902617 EDV43816.1 CH480819 EDW53410.1 CH954182 EDV53178.1 CM000160 EDW98247.1 CH916373 EDV94387.1 EZ422607 ADD18883.1 CM000070 EAL28122.2 OUUW01000005 SPP81053.1 GDAI01001943 JAI15660.1 JXJN01007738 CH479179 EDW24200.1 NNAY01000193 OXU30112.1 GEDC01005861 JAS31437.1 GEDC01005866 JAS31432.1 KQ981285 KYN43203.1 CP012526 ALC46919.1 GL447336 EFN86540.1 GL888087 EGI67719.1 QOIP01000007 RLU20958.1 GAMD01002514 JAA99076.1 KK107364 EZA51931.1 KQ980832 KYN12396.1 GDIP01101655 JAM02060.1 KQ414667 KOC64741.1 JXUM01042943 KQ561344 KXJ78885.1 KZ288194 PBC33942.1 KQ982289 KYQ58424.1 GL767232 EFZ13990.1 KQ978068 KYM97608.1 NEVH01006756 PNF36424.1 KQ435863 KOX70355.1 AMQN01010616 KB308243 ELT98080.1 GL732526 EFX87971.1 GL443112 EFN62570.1 CH964232 EDW81187.1 ADTU01017112 GDIP01186102 JAJ37300.1 GDIP01230888 LRGB01001361 JAI92513.1 KZS12361.1 GBRD01016656 GDHC01008634 JAG49171.1 JAQ09995.1 KQ976394 KYM93244.1 GBHO01028369 JAG15235.1 GDIP01148506 JAJ74896.1 GDIP01120255 JAL83459.1 CCAG010013281 GAMC01003537 GAMC01003535 JAC03019.1 AY652655 AAV67032.1 GEFM01003792 JAP72004.1 GDIP01107208 JAL96506.1 GADI01001197 JAA72611.1
Proteomes
UP000005204
UP000053268
UP000007151
UP000037510
UP000218220
UP000053240
+ More
UP000037069 UP000007266 UP000030765 UP000076408 UP000245037 UP000092461 UP000075880 UP000069272 UP000095301 UP000075920 UP000000673 UP000075886 UP000008820 UP000075903 UP000076407 UP000007062 UP000075900 UP000009046 UP000075884 UP000075882 UP000092462 UP000027135 UP000000803 UP000076502 UP000075881 UP000095300 UP000009192 UP000002320 UP000008792 UP000007801 UP000001292 UP000008711 UP000002282 UP000001070 UP000001819 UP000192223 UP000268350 UP000092460 UP000002358 UP000008744 UP000215335 UP000092443 UP000078541 UP000092553 UP000008237 UP000192221 UP000007755 UP000279307 UP000053097 UP000078492 UP000078200 UP000053825 UP000069940 UP000249989 UP000242457 UP000091820 UP000075809 UP000078542 UP000235965 UP000053105 UP000014760 UP000000305 UP000000311 UP000007798 UP000005205 UP000076858 UP000078540 UP000092445 UP000092444
UP000037069 UP000007266 UP000030765 UP000076408 UP000245037 UP000092461 UP000075880 UP000069272 UP000095301 UP000075920 UP000000673 UP000075886 UP000008820 UP000075903 UP000076407 UP000007062 UP000075900 UP000009046 UP000075884 UP000075882 UP000092462 UP000027135 UP000000803 UP000076502 UP000075881 UP000095300 UP000009192 UP000002320 UP000008792 UP000007801 UP000001292 UP000008711 UP000002282 UP000001070 UP000001819 UP000192223 UP000268350 UP000092460 UP000002358 UP000008744 UP000215335 UP000092443 UP000078541 UP000092553 UP000008237 UP000192221 UP000007755 UP000279307 UP000053097 UP000078492 UP000078200 UP000053825 UP000069940 UP000249989 UP000242457 UP000091820 UP000075809 UP000078542 UP000235965 UP000053105 UP000014760 UP000000305 UP000000311 UP000007798 UP000005205 UP000076858 UP000078540 UP000092445 UP000092444
Pfam
PF00459 Inositol_P
Interpro
ProteinModelPortal
H9J5X8
A0A194PG39
A0A2H1WLP3
A0A212ETS0
A0A0L7LEW9
A0A2A4JLR9
+ More
A0A194RQC8 A0A0L0BYC9 D6WQP1 A0A084W804 A0A182YEP8 A0A1S4F9I5 A0A2P8YXD2 A0A1B0CJ88 A0A182IXC6 A0A182F8R9 A0A2M4ALZ1 A0A1I8MRT0 A0A182VPX4 A0A2M4BV34 A0A1L8ECR1 W5JHP7 A0A182QXQ2 Q17B69 A0A182VB10 A0A182XRT0 Q7Q502 A0A182RE73 E0VSL4 A0A1L8DMG7 U5EYU6 A0A182NN51 A0A182LQ68 A0A1B0CYY1 A0A067QMS0 A0A1Q3FGU0 Q9VAG9 A0A154P4T9 A0A182JWZ2 A0A1Y1MF75 A0A1I8P0S4 B4KAN4 B0W9Q0 B4M0P9 B3LWX7 B4HZF3 B3P5C8 B4PPJ7 B4JRJ7 D3TMF5 Q297T3 A0A1W4XCE3 A0A3B0JJT5 A0A0K8TP35 A0A1B0B300 K7J1G5 B4G305 A0A232FHF8 A0A240SWM1 A0A1B6E0N3 A0A1B6E0G8 A0A195FRX8 A0A0M3QY31 E2BCN0 A0A1W4UR76 F4WDD9 A0A3L8DMB2 T1DQG2 A0A026W8F8 A0A151IX35 A0A1A9VDQ5 A0A0N8CT24 A0A0L7R1S5 A0A182G572 A0A2A3ERP2 A0A1A9W5P8 A0A151XDH1 E9IZR7 A0A195CBZ0 A0A2J7R6G7 A0A0M8ZSQ1 R7U4I7 E9FWX6 E2AVH5 B4NBM3 A0A158NIS7 A0A0P5BE40 A0A0P4YHI5 A0A0K8S789 A0A195BX77 A0A1A9ZSU8 A0A0A9XDR7 A0A0P5E012 A0A0N8CLE9 A0A1B0FLB9 W8C1J4 Q4VRW1 A0A131XXM0 A0A0P5UZF2 A0A0K8RNI9
A0A194RQC8 A0A0L0BYC9 D6WQP1 A0A084W804 A0A182YEP8 A0A1S4F9I5 A0A2P8YXD2 A0A1B0CJ88 A0A182IXC6 A0A182F8R9 A0A2M4ALZ1 A0A1I8MRT0 A0A182VPX4 A0A2M4BV34 A0A1L8ECR1 W5JHP7 A0A182QXQ2 Q17B69 A0A182VB10 A0A182XRT0 Q7Q502 A0A182RE73 E0VSL4 A0A1L8DMG7 U5EYU6 A0A182NN51 A0A182LQ68 A0A1B0CYY1 A0A067QMS0 A0A1Q3FGU0 Q9VAG9 A0A154P4T9 A0A182JWZ2 A0A1Y1MF75 A0A1I8P0S4 B4KAN4 B0W9Q0 B4M0P9 B3LWX7 B4HZF3 B3P5C8 B4PPJ7 B4JRJ7 D3TMF5 Q297T3 A0A1W4XCE3 A0A3B0JJT5 A0A0K8TP35 A0A1B0B300 K7J1G5 B4G305 A0A232FHF8 A0A240SWM1 A0A1B6E0N3 A0A1B6E0G8 A0A195FRX8 A0A0M3QY31 E2BCN0 A0A1W4UR76 F4WDD9 A0A3L8DMB2 T1DQG2 A0A026W8F8 A0A151IX35 A0A1A9VDQ5 A0A0N8CT24 A0A0L7R1S5 A0A182G572 A0A2A3ERP2 A0A1A9W5P8 A0A151XDH1 E9IZR7 A0A195CBZ0 A0A2J7R6G7 A0A0M8ZSQ1 R7U4I7 E9FWX6 E2AVH5 B4NBM3 A0A158NIS7 A0A0P5BE40 A0A0P4YHI5 A0A0K8S789 A0A195BX77 A0A1A9ZSU8 A0A0A9XDR7 A0A0P5E012 A0A0N8CLE9 A0A1B0FLB9 W8C1J4 Q4VRW1 A0A131XXM0 A0A0P5UZF2 A0A0K8RNI9
PDB
2WEF
E-value=1.52304e-81,
Score=770
Ontologies
PATHWAY
GO
Topology
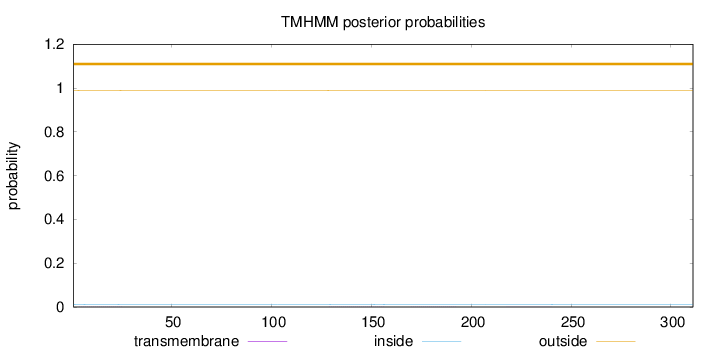
Length:
311
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.04637
Exp number, first 60 AAs:
0.01362
Total prob of N-in:
0.01202
outside
1 - 311
Population Genetic Test Statistics
Pi
212.143141
Theta
175.282292
Tajima's D
0.703953
CLR
0.356956
CSRT
0.567671616419179
Interpretation
Uncertain
