Gene
KWMTBOMO15128
Annotation
hypothetical_protein_KGM_11337_[Danaus_plexippus]
Location in the cell
Mitochondrial Reliability : 1.495 Nuclear Reliability : 1.195
Sequence
CDS
ATGGGTAGCCTTGACAGAGCAATCCTAACTGGGTTCATATGTCGACTATGTTCTGAAATGCACCGCATTGTGATACATATATATGGTGAAGAAGGCATACGACTATGTATTTCAGAAAAAATCAGTCGATATCTCACCATAAACATTTCTCGAGCCGACCCACTCCCAAAAACCATTTGCAAAAATTGTTTGGAAAGACTTGAAAAACAACACAAACTGGTAATGGTGATGGAAAACGCAGCCAACATGTTAAAAGGGCGTAAAACAAGGGCTGCCAAAAGTGAAACAAAGCAGTAA
Protein
MGSLDRAILTGFICRLCSEMHRIVIHIYGEEGIRLCISEKISRYLTINISRADPLPKTICKNCLERLEKQHKLVMVMENAANMLKGRKTRAAKSETKQ
Summary
Uniprot
A0A212ETS4
S4Q0A1
A0A3S2P1M9
A0A2A4J2C3
A0A2H1W1R3
A0A0L7LEU5
+ More
A0A194RLD8 A0A0C9R3W7 A0A0K8V8Z8 B4NIX5 A0A1A9YT48 A0A1B0B3E9 A0A1I8PND7 A0A1A9UVH3 A0A0K8UAK5 A0A0K8U9N9 A0A1A9WYQ8 A0A1I8MQL3 A0A0A1WW20 A0A1B6IXS0 A0A0A1XNG3 B4K4H0 B4JYT9 A0A0Q9WKK8 B4MCA1 Q9VBY9 B3P6T4 A0A1W4VLS4 B4IJE4 B4QU98 B4PUF1 A0A0N1ITV8 F4WC42 A0A195BJ11 A0A195FRJ0 E2A2D5 A0A2A3EBF7 A0A087ZTC8 A0A0L7QMD4 Q297F9 B4GE41 A0A3L8D9N1 A0A0J7KVS4 A0A154PJA4 A0A336M609 A0A158NAQ6 E2BZ10 A0A2P8YW52 T1GGL5 A0A067QIC2 B3M1Y0 A0A232F7W8 K7IQD1 A0A026WLE6 A0A3Q0IIB4 A0A0K8TN54 A0A151WX10 A0A195CWA7 A0A224Y1K5 A0A0P4VLD0 A0A0V0GDF6 A0A069DNQ0 E0VIF9 A0A2R7WBR1 A0A195EKE5 R4WKH4 A0A182H6Y8 A0A023EJ94 Q17CI4 A0A1Q3G220 A0A1J1I2W8 A0A0A9WMR0 A0A1B0CB29 A0A084WQT0 A0A182RNG9 A0A182VR90 A0A1L8D8E1 A0A182YGG0 A0A1Y9H9F4 A0A182X9T6 A0A182L919 Q7PXH0 A0A182HKR8 A0A182VLK4 J9KWU5 A0A182P2G8 A0A0K8UF42 D6WZB2 A0A182N5D5 A0A182S758 V5G8E1 A0A182F797 W5JJ64 A0A1W4XNP1 A0A2M4AAV9 B4H0E9 A0A1Y1MJM6
A0A194RLD8 A0A0C9R3W7 A0A0K8V8Z8 B4NIX5 A0A1A9YT48 A0A1B0B3E9 A0A1I8PND7 A0A1A9UVH3 A0A0K8UAK5 A0A0K8U9N9 A0A1A9WYQ8 A0A1I8MQL3 A0A0A1WW20 A0A1B6IXS0 A0A0A1XNG3 B4K4H0 B4JYT9 A0A0Q9WKK8 B4MCA1 Q9VBY9 B3P6T4 A0A1W4VLS4 B4IJE4 B4QU98 B4PUF1 A0A0N1ITV8 F4WC42 A0A195BJ11 A0A195FRJ0 E2A2D5 A0A2A3EBF7 A0A087ZTC8 A0A0L7QMD4 Q297F9 B4GE41 A0A3L8D9N1 A0A0J7KVS4 A0A154PJA4 A0A336M609 A0A158NAQ6 E2BZ10 A0A2P8YW52 T1GGL5 A0A067QIC2 B3M1Y0 A0A232F7W8 K7IQD1 A0A026WLE6 A0A3Q0IIB4 A0A0K8TN54 A0A151WX10 A0A195CWA7 A0A224Y1K5 A0A0P4VLD0 A0A0V0GDF6 A0A069DNQ0 E0VIF9 A0A2R7WBR1 A0A195EKE5 R4WKH4 A0A182H6Y8 A0A023EJ94 Q17CI4 A0A1Q3G220 A0A1J1I2W8 A0A0A9WMR0 A0A1B0CB29 A0A084WQT0 A0A182RNG9 A0A182VR90 A0A1L8D8E1 A0A182YGG0 A0A1Y9H9F4 A0A182X9T6 A0A182L919 Q7PXH0 A0A182HKR8 A0A182VLK4 J9KWU5 A0A182P2G8 A0A0K8UF42 D6WZB2 A0A182N5D5 A0A182S758 V5G8E1 A0A182F797 W5JJ64 A0A1W4XNP1 A0A2M4AAV9 B4H0E9 A0A1Y1MJM6
Pubmed
22118469
23622113
26227816
26354079
17994087
25315136
+ More
25830018 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 21719571 20798317 15632085 30249741 21347285 29403074 24845553 28648823 20075255 24508170 26369729 27129103 26334808 20566863 23691247 26483478 24945155 17510324 25401762 26823975 24438588 25244985 20966253 12364791 14747013 17210077 18362917 19820115 20920257 23761445 28004739
25830018 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 21719571 20798317 15632085 30249741 21347285 29403074 24845553 28648823 20075255 24508170 26369729 27129103 26334808 20566863 23691247 26483478 24945155 17510324 25401762 26823975 24438588 25244985 20966253 12364791 14747013 17210077 18362917 19820115 20920257 23761445 28004739
EMBL
AGBW02012531
OWR44893.1
GAIX01000148
JAA92412.1
RSAL01000053
RVE50070.1
+ More
NWSH01003479 PCG66215.1 ODYU01005792 SOQ47035.1 JTDY01001414 KOB73925.1 KQ460045 KPJ18234.1 GBYB01007497 JAG77264.1 GDHF01016962 JAI35352.1 CH964272 EDW84877.1 JXJN01007813 GDHF01028650 GDHF01021722 JAI23664.1 JAI30592.1 GDHF01029043 JAI23271.1 GBXI01011654 JAD02638.1 GECU01022366 GECU01015982 GECU01012610 JAS85340.1 JAS91724.1 JAS95096.1 GBXI01001795 JAD12497.1 CH933806 EDW13922.1 KRG00721.1 CH916378 EDV98554.1 CH940657 KRF80944.1 EDW63183.2 AE014297 AY071126 AAF56387.2 AAL48748.1 CH954182 EDV53754.1 CH480848 EDW51135.1 CM000364 EDX14335.1 CM000160 EDW98838.1 KQ435736 KOX77032.1 GL888070 EGI68022.1 KQ976455 KYM85086.1 KQ981305 KYN42902.1 GL436037 EFN72398.1 KZ288292 PBC29103.1 KQ414894 KOC59714.1 CM000070 EAL28246.3 CH479182 EDW33876.1 QOIP01000011 RLU17215.1 LBMM01002763 KMQ94403.1 KQ434905 KZC11280.1 UFQS01000609 UFQT01000609 SSX05335.1 SSX25696.1 ADTU01009943 GL451538 EFN79082.1 PYGN01000325 PSN48478.1 CAQQ02045615 KK853567 KDR06669.1 CH902617 EDV42240.1 NNAY01000779 OXU26548.1 KK107182 EZA55924.1 GDAI01001801 JAI15802.1 KQ982686 KYQ52297.1 KQ977276 KYN04439.1 GFTR01001534 JAW14892.1 GDKW01003375 JAI53220.1 GECL01000749 JAP05375.1 GBGD01003603 JAC85286.1 DS235199 EEB13165.1 KK854569 PTY16959.1 KQ978739 KYN28713.1 AK418090 BAN21305.1 JXUM01115307 KQ565867 KXJ70587.1 JXUM01143237 GAPW01004571 KQ569558 JAC09027.1 KXJ68594.1 CH477308 EAT44037.1 GFDL01001224 JAV33821.1 CVRI01000039 CRK94629.1 GBHO01035817 GBHO01035811 GDHC01000622 JAG07787.1 JAG07793.1 JAQ18007.1 AJWK01004781 ATLV01025723 KE525398 KFB52574.1 GFDF01011351 JAV02733.1 AXCN02001179 AAAB01008987 EAA01506.4 APCN01000860 ABLF02036116 ABLF02036118 GDHF01027070 JAI25244.1 KQ971372 EFA10399.1 GALX01008187 JAB60279.1 ADMH02001070 ETN64186.1 GGFK01004602 MBW37923.1 CH479200 EDW29744.1 GEZM01029807 JAV85914.1
NWSH01003479 PCG66215.1 ODYU01005792 SOQ47035.1 JTDY01001414 KOB73925.1 KQ460045 KPJ18234.1 GBYB01007497 JAG77264.1 GDHF01016962 JAI35352.1 CH964272 EDW84877.1 JXJN01007813 GDHF01028650 GDHF01021722 JAI23664.1 JAI30592.1 GDHF01029043 JAI23271.1 GBXI01011654 JAD02638.1 GECU01022366 GECU01015982 GECU01012610 JAS85340.1 JAS91724.1 JAS95096.1 GBXI01001795 JAD12497.1 CH933806 EDW13922.1 KRG00721.1 CH916378 EDV98554.1 CH940657 KRF80944.1 EDW63183.2 AE014297 AY071126 AAF56387.2 AAL48748.1 CH954182 EDV53754.1 CH480848 EDW51135.1 CM000364 EDX14335.1 CM000160 EDW98838.1 KQ435736 KOX77032.1 GL888070 EGI68022.1 KQ976455 KYM85086.1 KQ981305 KYN42902.1 GL436037 EFN72398.1 KZ288292 PBC29103.1 KQ414894 KOC59714.1 CM000070 EAL28246.3 CH479182 EDW33876.1 QOIP01000011 RLU17215.1 LBMM01002763 KMQ94403.1 KQ434905 KZC11280.1 UFQS01000609 UFQT01000609 SSX05335.1 SSX25696.1 ADTU01009943 GL451538 EFN79082.1 PYGN01000325 PSN48478.1 CAQQ02045615 KK853567 KDR06669.1 CH902617 EDV42240.1 NNAY01000779 OXU26548.1 KK107182 EZA55924.1 GDAI01001801 JAI15802.1 KQ982686 KYQ52297.1 KQ977276 KYN04439.1 GFTR01001534 JAW14892.1 GDKW01003375 JAI53220.1 GECL01000749 JAP05375.1 GBGD01003603 JAC85286.1 DS235199 EEB13165.1 KK854569 PTY16959.1 KQ978739 KYN28713.1 AK418090 BAN21305.1 JXUM01115307 KQ565867 KXJ70587.1 JXUM01143237 GAPW01004571 KQ569558 JAC09027.1 KXJ68594.1 CH477308 EAT44037.1 GFDL01001224 JAV33821.1 CVRI01000039 CRK94629.1 GBHO01035817 GBHO01035811 GDHC01000622 JAG07787.1 JAG07793.1 JAQ18007.1 AJWK01004781 ATLV01025723 KE525398 KFB52574.1 GFDF01011351 JAV02733.1 AXCN02001179 AAAB01008987 EAA01506.4 APCN01000860 ABLF02036116 ABLF02036118 GDHF01027070 JAI25244.1 KQ971372 EFA10399.1 GALX01008187 JAB60279.1 ADMH02001070 ETN64186.1 GGFK01004602 MBW37923.1 CH479200 EDW29744.1 GEZM01029807 JAV85914.1
Proteomes
UP000007151
UP000283053
UP000218220
UP000037510
UP000053240
UP000007798
+ More
UP000092443 UP000092460 UP000095300 UP000078200 UP000091820 UP000095301 UP000009192 UP000001070 UP000008792 UP000000803 UP000008711 UP000192221 UP000001292 UP000000304 UP000002282 UP000053105 UP000007755 UP000078540 UP000078541 UP000000311 UP000242457 UP000005203 UP000053825 UP000001819 UP000008744 UP000279307 UP000036403 UP000076502 UP000005205 UP000008237 UP000245037 UP000015102 UP000027135 UP000007801 UP000215335 UP000002358 UP000053097 UP000079169 UP000075809 UP000078542 UP000009046 UP000078492 UP000069940 UP000249989 UP000008820 UP000183832 UP000092461 UP000030765 UP000075900 UP000075920 UP000076408 UP000075886 UP000076407 UP000075882 UP000007062 UP000075840 UP000075903 UP000007819 UP000075885 UP000007266 UP000075884 UP000075901 UP000069272 UP000000673 UP000192223
UP000092443 UP000092460 UP000095300 UP000078200 UP000091820 UP000095301 UP000009192 UP000001070 UP000008792 UP000000803 UP000008711 UP000192221 UP000001292 UP000000304 UP000002282 UP000053105 UP000007755 UP000078540 UP000078541 UP000000311 UP000242457 UP000005203 UP000053825 UP000001819 UP000008744 UP000279307 UP000036403 UP000076502 UP000005205 UP000008237 UP000245037 UP000015102 UP000027135 UP000007801 UP000215335 UP000002358 UP000053097 UP000079169 UP000075809 UP000078542 UP000009046 UP000078492 UP000069940 UP000249989 UP000008820 UP000183832 UP000092461 UP000030765 UP000075900 UP000075920 UP000076408 UP000075886 UP000076407 UP000075882 UP000007062 UP000075840 UP000075903 UP000007819 UP000075885 UP000007266 UP000075884 UP000075901 UP000069272 UP000000673 UP000192223
PRIDE
Pfam
PF07776 zf-AD
Interpro
IPR012934
Znf_AD
ProteinModelPortal
A0A212ETS4
S4Q0A1
A0A3S2P1M9
A0A2A4J2C3
A0A2H1W1R3
A0A0L7LEU5
+ More
A0A194RLD8 A0A0C9R3W7 A0A0K8V8Z8 B4NIX5 A0A1A9YT48 A0A1B0B3E9 A0A1I8PND7 A0A1A9UVH3 A0A0K8UAK5 A0A0K8U9N9 A0A1A9WYQ8 A0A1I8MQL3 A0A0A1WW20 A0A1B6IXS0 A0A0A1XNG3 B4K4H0 B4JYT9 A0A0Q9WKK8 B4MCA1 Q9VBY9 B3P6T4 A0A1W4VLS4 B4IJE4 B4QU98 B4PUF1 A0A0N1ITV8 F4WC42 A0A195BJ11 A0A195FRJ0 E2A2D5 A0A2A3EBF7 A0A087ZTC8 A0A0L7QMD4 Q297F9 B4GE41 A0A3L8D9N1 A0A0J7KVS4 A0A154PJA4 A0A336M609 A0A158NAQ6 E2BZ10 A0A2P8YW52 T1GGL5 A0A067QIC2 B3M1Y0 A0A232F7W8 K7IQD1 A0A026WLE6 A0A3Q0IIB4 A0A0K8TN54 A0A151WX10 A0A195CWA7 A0A224Y1K5 A0A0P4VLD0 A0A0V0GDF6 A0A069DNQ0 E0VIF9 A0A2R7WBR1 A0A195EKE5 R4WKH4 A0A182H6Y8 A0A023EJ94 Q17CI4 A0A1Q3G220 A0A1J1I2W8 A0A0A9WMR0 A0A1B0CB29 A0A084WQT0 A0A182RNG9 A0A182VR90 A0A1L8D8E1 A0A182YGG0 A0A1Y9H9F4 A0A182X9T6 A0A182L919 Q7PXH0 A0A182HKR8 A0A182VLK4 J9KWU5 A0A182P2G8 A0A0K8UF42 D6WZB2 A0A182N5D5 A0A182S758 V5G8E1 A0A182F797 W5JJ64 A0A1W4XNP1 A0A2M4AAV9 B4H0E9 A0A1Y1MJM6
A0A194RLD8 A0A0C9R3W7 A0A0K8V8Z8 B4NIX5 A0A1A9YT48 A0A1B0B3E9 A0A1I8PND7 A0A1A9UVH3 A0A0K8UAK5 A0A0K8U9N9 A0A1A9WYQ8 A0A1I8MQL3 A0A0A1WW20 A0A1B6IXS0 A0A0A1XNG3 B4K4H0 B4JYT9 A0A0Q9WKK8 B4MCA1 Q9VBY9 B3P6T4 A0A1W4VLS4 B4IJE4 B4QU98 B4PUF1 A0A0N1ITV8 F4WC42 A0A195BJ11 A0A195FRJ0 E2A2D5 A0A2A3EBF7 A0A087ZTC8 A0A0L7QMD4 Q297F9 B4GE41 A0A3L8D9N1 A0A0J7KVS4 A0A154PJA4 A0A336M609 A0A158NAQ6 E2BZ10 A0A2P8YW52 T1GGL5 A0A067QIC2 B3M1Y0 A0A232F7W8 K7IQD1 A0A026WLE6 A0A3Q0IIB4 A0A0K8TN54 A0A151WX10 A0A195CWA7 A0A224Y1K5 A0A0P4VLD0 A0A0V0GDF6 A0A069DNQ0 E0VIF9 A0A2R7WBR1 A0A195EKE5 R4WKH4 A0A182H6Y8 A0A023EJ94 Q17CI4 A0A1Q3G220 A0A1J1I2W8 A0A0A9WMR0 A0A1B0CB29 A0A084WQT0 A0A182RNG9 A0A182VR90 A0A1L8D8E1 A0A182YGG0 A0A1Y9H9F4 A0A182X9T6 A0A182L919 Q7PXH0 A0A182HKR8 A0A182VLK4 J9KWU5 A0A182P2G8 A0A0K8UF42 D6WZB2 A0A182N5D5 A0A182S758 V5G8E1 A0A182F797 W5JJ64 A0A1W4XNP1 A0A2M4AAV9 B4H0E9 A0A1Y1MJM6
Ontologies
GO
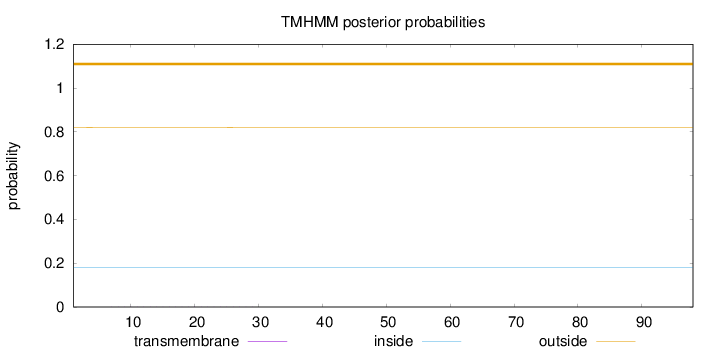
Topology
Length:
98
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01055
Exp number, first 60 AAs:
0.01055
Total prob of N-in:
0.17940
outside
1 - 98
Population Genetic Test Statistics
Pi
66.824558
Theta
124.283085
Tajima's D
-1.16834
CLR
0
CSRT
0.111094445277736
Interpretation
Uncertain
