Gene
KWMTBOMO15123 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA004908
Annotation
PREDICTED:_26S_protease_regulatory_subunit_4_[Bombyx_mori]
Full name
26S proteasome regulatory subunit 4
Location in the cell
Cytoplasmic Reliability : 3.012
Sequence
CDS
ATGGGTCAAAATCAATCAGGTGGAGGTGGTGACAAAAAAGATGACAAAGACAAGAAGAAGAAGTATGAGCCTCCGATCCCTACACGAGTTGGTAAAAAGAAAAGGAAAGCTAAGGGACCAGATGCGGCTCTCAAGCTTCCGCAAGTCACGCCTCATACAAGATGTCGACTCAAGTTGTTAAAACTCGAAAGAATAAAAGACTATTTGTTGATGGAAGAGGAGTTCATCCGTAATCAAGAACGCTTGAAGCCACAGGAAGAGAAAATTGAAGAAGAAAGGTCTAAGGTTGATGACCTAAGAGGAACTCCAATGTCAGTTGGTAATCTTGAGGAAATCATAGATGACAATCATGCAATAGTGTCAACTTCAGTTGGCAGTGAACATTATGTTAGCATTCTCTCCTTTGTTGACAAGGATCAGCTTGAACCTGGATGCTCTGTGTTACTGAATCATAAGGTGCACGCAGTTGTTGGTGTTTTGGGTGATGACACGGATCCCATGGTGTCAGTCATGAAGCTTGAAAAGGCTCCACAGGAAACCTATGCTGACATTGGTGGCCTGGACACCCAGATCCAGGAAATCAAGGAATCTGTGGAGTTGCCTCTAACACATCCGGAATACTATGAAGAAATGGGAATCAAACCTCCCAAAGGAGTCATCTTGTACGGTCCTCCGGGCACTGGTAAAACACTCCTGGCCAAGGCTGTTGCAAATCAGACATCTGCTACCTTCCTACGTGTAGTTGGATCTGAACTTATACAAAAATATTTGGGTGATGGTCCCAAGTTGGTTCGTGAACTGTTCCGTGTTGCTGAGGAACATGCACCTTCAATTGTTTTCATTGATGAAATTGATGCAGTGGGCACTAAACGTTATGATTCAAACTCTGGTGGTGAAAGGGAAATTCAAAGGACCATGTTGGAATTATTGAATCAACTTGATGGTTTTGATTCAAGAGGAGATGTGAAGGTGATCATGGCCACAAACCGTATAGAAACTTTAGACCCGGCACTTATCCGACCAGGTCGTATTGATAGGAAGATTGAATTCCCCTTGCCTGACGAGAAAACCAAGAGAAGGATTTTCACCATTCATACATCAAGAATGACATTAGCTGATGATGTGAACTTGTCTGAACTTATTATGTCAAAAGATGATCTTTCTGGTGCTGATATTAAGGCAATATGTACTGAAGCTGGTCTCATGGCACTTAGAGAGCGCCGTATGAAAGTAACAAATGAGGACTTCAAGAAATCAAAGGAAAGTGTCCTATACCGCAAGAAGGAAGGCACACCAGAAGGACTTTATTTATAA
Protein
MGQNQSGGGGDKKDDKDKKKKYEPPIPTRVGKKKRKAKGPDAALKLPQVTPHTRCRLKLLKLERIKDYLLMEEEFIRNQERLKPQEEKIEEERSKVDDLRGTPMSVGNLEEIIDDNHAIVSTSVGSEHYVSILSFVDKDQLEPGCSVLLNHKVHAVVGVLGDDTDPMVSVMKLEKAPQETYADIGGLDTQIQEIKESVELPLTHPEYYEEMGIKPPKGVILYGPPGTGKTLLAKAVANQTSATFLRVVGSELIQKYLGDGPKLVRELFRVAEEHAPSIVFIDEIDAVGTKRYDSNSGGEREIQRTMLELLNQLDGFDSRGDVKVIMATNRIETLDPALIRPGRIDRKIEFPLPDEKTKRRIFTIHTSRMTLADDVNLSELIMSKDDLSGADIKAICTEAGLMALRERRMKVTNEDFKKSKESVLYRKKEGTPEGLYL
Summary
Description
The 26S proteasome is involved in the ATP-dependent degradation of ubiquitinated proteins. The regulatory (or ATPase) complex confers ATP dependency and substrate specificity to the 26S complex (By similarity).
Subunit
Interacts with PSMD5.
Similarity
Belongs to the AAA ATPase family.
Belongs to the UDP-glycosyltransferase family.
Belongs to the UDP-glycosyltransferase family.
Keywords
ATP-binding
Complete proteome
Cytoplasm
Nucleotide-binding
Nucleus
Proteasome
Reference proteome
Feature
chain 26S proteasome regulatory subunit 4
Uniprot
A0A2A4JHF8
A0A3S2NVS4
A0A2H1W1T9
A0A194PG29
S4PC80
A0A182FSK9
+ More
I4DND5 A0A1J1HPP4 A0A182QB88 T1E2M9 A0A2M3Z2A7 A0A2M4A938 A0A2M4BNF7 A0A182PE42 E3WN61 A0A182N5B1 B0WWV3 A0A182TUL9 A0A182XE26 A0A182LCH1 A0T4G5 A0A182HJ96 Q1HQY1 A0A023ES93 A0A1W4UK15 B4QSD0 A0A1Q3FDQ6 A0A0B4LIJ0 B4PL52 B3M322 B4NBQ9 B4HG06 B3P8C1 P48601 A0A182G9P6 A0A1L8DT25 A0A0L0BMZ9 B4JRP6 B4M0C9 A0A3B0KCN1 A0A084WSL1 D3TMJ5 A0A1A9UQ60 A0A1B0BZ88 Q298N1 A0A1L8EHU8 A0A1B0A8W2 A0A1A9W6H4 U5EX60 A0A182XUX0 A0A067RGZ7 A0A336LG10 B4K736 A0A1I8QEI4 W8B0V2 E2BWH6 T1P826 A0A069DUE0 R4FNS2 A0A1W4X6M8 A0A0M4EK14 J3JXA9 A0A0K8UH06 A0A034W729 A0A2J7R280 R4WD80 A0A0B4J2L7 A0A182V0H7 A0A224XNL3 A0A0L7R1J4 V9IG44 A0A1B6HWI1 D6WTT2 A0A2A3EQB0 A0A0C9RQW5 A0A2R7VYI9 A0A026W6M8 A0A0A9WW95 A0A1B6MFA0 A0A1B6DDK7 A0A1Y1KB25 F4W4Q7 A0A0J7L139 E2ALJ9 A0A232ETH9 A0A0P4VME7 T1JN31 V5G8R3 E0W054 K7J1D7 A0A0P4WS62 A0A0P5GTX9 A0A2P6KDC2 A0A226ESC3 A0A3S1BJS8 A0A1D2MMR6 A0A1B0FHF8 A0A0K8TN88 A0A0B7BB55 C3XS01
I4DND5 A0A1J1HPP4 A0A182QB88 T1E2M9 A0A2M3Z2A7 A0A2M4A938 A0A2M4BNF7 A0A182PE42 E3WN61 A0A182N5B1 B0WWV3 A0A182TUL9 A0A182XE26 A0A182LCH1 A0T4G5 A0A182HJ96 Q1HQY1 A0A023ES93 A0A1W4UK15 B4QSD0 A0A1Q3FDQ6 A0A0B4LIJ0 B4PL52 B3M322 B4NBQ9 B4HG06 B3P8C1 P48601 A0A182G9P6 A0A1L8DT25 A0A0L0BMZ9 B4JRP6 B4M0C9 A0A3B0KCN1 A0A084WSL1 D3TMJ5 A0A1A9UQ60 A0A1B0BZ88 Q298N1 A0A1L8EHU8 A0A1B0A8W2 A0A1A9W6H4 U5EX60 A0A182XUX0 A0A067RGZ7 A0A336LG10 B4K736 A0A1I8QEI4 W8B0V2 E2BWH6 T1P826 A0A069DUE0 R4FNS2 A0A1W4X6M8 A0A0M4EK14 J3JXA9 A0A0K8UH06 A0A034W729 A0A2J7R280 R4WD80 A0A0B4J2L7 A0A182V0H7 A0A224XNL3 A0A0L7R1J4 V9IG44 A0A1B6HWI1 D6WTT2 A0A2A3EQB0 A0A0C9RQW5 A0A2R7VYI9 A0A026W6M8 A0A0A9WW95 A0A1B6MFA0 A0A1B6DDK7 A0A1Y1KB25 F4W4Q7 A0A0J7L139 E2ALJ9 A0A232ETH9 A0A0P4VME7 T1JN31 V5G8R3 E0W054 K7J1D7 A0A0P4WS62 A0A0P5GTX9 A0A2P6KDC2 A0A226ESC3 A0A3S1BJS8 A0A1D2MMR6 A0A1B0FHF8 A0A0K8TN88 A0A0B7BB55 C3XS01
Pubmed
26354079
23622113
22651552
24330624
20920257
23761445
+ More
20966253 12364791 14747013 17210077 17204158 17510324 24945155 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 8808288 12537569 23622245 26483478 26108605 24438588 20353571 15632085 25244985 24845553 24495485 20798317 25315136 26334808 22516182 23537049 25348373 23691247 18362917 19820115 24508170 25401762 26823975 28004739 21719571 28648823 27129103 20566863 20075255 27289101 26369729 18563158
20966253 12364791 14747013 17210077 17204158 17510324 24945155 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 8808288 12537569 23622245 26483478 26108605 24438588 20353571 15632085 25244985 24845553 24495485 20798317 25315136 26334808 22516182 23537049 25348373 23691247 18362917 19820115 24508170 25401762 26823975 28004739 21719571 28648823 27129103 20566863 20075255 27289101 26369729 18563158
EMBL
NWSH01001392
PCG71411.1
RSAL01000053
RVE50066.1
ODYU01005792
SOQ47028.1
+ More
KQ459604 KPI92252.1 GAIX01005582 JAA86978.1 AK402858 BAM19425.1 CVRI01000008 CRK88438.1 AXCN02000148 GALA01001135 JAA93717.1 GGFM01001908 MBW22659.1 GGFK01003964 MBW37285.1 GGFJ01005444 MBW54585.1 ADMH02002074 ETN59541.1 ETN59542.1 DS232152 EDS36214.1 AAAB01008879 EAA08386.4 EAA08387.4 APCN01002133 DQ440313 CH477841 ABF18346.1 EAT35749.1 GAPW01001486 JAC12112.1 CM000364 EDX14129.1 GFDL01009344 JAV25701.1 AE014297 AHN57492.1 CM000160 EDW99037.1 CH902617 EDV42422.2 CH964232 EDW81223.1 CH480815 EDW43399.1 CH954182 EDV53945.1 U39303 AY058759 JXUM01049616 KQ561613 KXJ78005.1 GFDF01004560 JAV09524.1 JRES01001623 KNC21396.1 CH916373 EDV94436.1 CH940650 EDW67291.2 OUUW01000007 SPP82771.1 ATLV01026615 KE525415 KFB53205.1 EZ422647 ADD18923.1 JXJN01023016 CM000070 EAL27924.1 GFDG01000575 JAV18224.1 GANO01001108 JAB58763.1 KK852643 KDR19574.1 UFQS01003326 UFQT01003326 SSX15505.1 SSX34871.1 CH933806 EDW16349.1 GAMC01019655 GAMC01019649 GAMC01019644 GAMC01019640 JAB86911.1 GL451131 EFN79932.1 KA644699 AFP59328.1 GBGD01001493 JAC87396.1 ACPB03013520 GAHY01000463 JAA77047.1 CP012526 ALC47421.1 APGK01051054 BT127878 KB741181 AEE62840.1 ENN73080.1 GDHF01026468 JAI25846.1 GAKP01007581 GAKP01007576 GAKP01007575 GAKP01007572 JAC51376.1 NEVH01008200 PNF34943.1 AK417299 BAN20514.1 GFTR01006254 JAW10172.1 KQ414667 KOC64717.1 JR040801 AEY59304.1 GECU01028683 JAS79023.1 KQ971352 EFA06741.1 KZ288194 PBC33918.1 GBYB01010915 GBYB01010916 JAG80682.1 JAG80683.1 KK854173 PTY12573.1 KK107405 EZA51286.1 GBHO01030862 GBRD01016386 GDHC01001057 JAG12742.1 JAG49440.1 JAQ17572.1 GEBQ01005374 JAT34603.1 GEDC01013539 JAS23759.1 GEZM01089123 JAV57678.1 GL887547 EGI70823.1 LBMM01001445 KMQ96298.1 GL440609 EFN65701.1 NNAY01002276 OXU21652.1 GDKW01000889 JAI55706.1 JH431265 GALX01001961 JAB66505.1 DS235857 EEB19010.1 GDRN01035851 JAI67379.1 GDIQ01239804 JAK11921.1 MWRG01015336 PRD24323.1 LNIX01000002 OXA59486.1 RQTK01000172 RUS85375.1 LJIJ01000839 ODM94222.1 CCAG010022398 GDAI01002203 JAI15400.1 HACG01042636 CEK89501.1 GG666456 EEN69469.1
KQ459604 KPI92252.1 GAIX01005582 JAA86978.1 AK402858 BAM19425.1 CVRI01000008 CRK88438.1 AXCN02000148 GALA01001135 JAA93717.1 GGFM01001908 MBW22659.1 GGFK01003964 MBW37285.1 GGFJ01005444 MBW54585.1 ADMH02002074 ETN59541.1 ETN59542.1 DS232152 EDS36214.1 AAAB01008879 EAA08386.4 EAA08387.4 APCN01002133 DQ440313 CH477841 ABF18346.1 EAT35749.1 GAPW01001486 JAC12112.1 CM000364 EDX14129.1 GFDL01009344 JAV25701.1 AE014297 AHN57492.1 CM000160 EDW99037.1 CH902617 EDV42422.2 CH964232 EDW81223.1 CH480815 EDW43399.1 CH954182 EDV53945.1 U39303 AY058759 JXUM01049616 KQ561613 KXJ78005.1 GFDF01004560 JAV09524.1 JRES01001623 KNC21396.1 CH916373 EDV94436.1 CH940650 EDW67291.2 OUUW01000007 SPP82771.1 ATLV01026615 KE525415 KFB53205.1 EZ422647 ADD18923.1 JXJN01023016 CM000070 EAL27924.1 GFDG01000575 JAV18224.1 GANO01001108 JAB58763.1 KK852643 KDR19574.1 UFQS01003326 UFQT01003326 SSX15505.1 SSX34871.1 CH933806 EDW16349.1 GAMC01019655 GAMC01019649 GAMC01019644 GAMC01019640 JAB86911.1 GL451131 EFN79932.1 KA644699 AFP59328.1 GBGD01001493 JAC87396.1 ACPB03013520 GAHY01000463 JAA77047.1 CP012526 ALC47421.1 APGK01051054 BT127878 KB741181 AEE62840.1 ENN73080.1 GDHF01026468 JAI25846.1 GAKP01007581 GAKP01007576 GAKP01007575 GAKP01007572 JAC51376.1 NEVH01008200 PNF34943.1 AK417299 BAN20514.1 GFTR01006254 JAW10172.1 KQ414667 KOC64717.1 JR040801 AEY59304.1 GECU01028683 JAS79023.1 KQ971352 EFA06741.1 KZ288194 PBC33918.1 GBYB01010915 GBYB01010916 JAG80682.1 JAG80683.1 KK854173 PTY12573.1 KK107405 EZA51286.1 GBHO01030862 GBRD01016386 GDHC01001057 JAG12742.1 JAG49440.1 JAQ17572.1 GEBQ01005374 JAT34603.1 GEDC01013539 JAS23759.1 GEZM01089123 JAV57678.1 GL887547 EGI70823.1 LBMM01001445 KMQ96298.1 GL440609 EFN65701.1 NNAY01002276 OXU21652.1 GDKW01000889 JAI55706.1 JH431265 GALX01001961 JAB66505.1 DS235857 EEB19010.1 GDRN01035851 JAI67379.1 GDIQ01239804 JAK11921.1 MWRG01015336 PRD24323.1 LNIX01000002 OXA59486.1 RQTK01000172 RUS85375.1 LJIJ01000839 ODM94222.1 CCAG010022398 GDAI01002203 JAI15400.1 HACG01042636 CEK89501.1 GG666456 EEN69469.1
Proteomes
UP000218220
UP000283053
UP000053268
UP000069272
UP000183832
UP000075886
+ More
UP000075885 UP000000673 UP000075884 UP000002320 UP000075902 UP000076407 UP000075882 UP000007062 UP000075840 UP000008820 UP000192221 UP000000304 UP000000803 UP000002282 UP000007801 UP000007798 UP000001292 UP000008711 UP000069940 UP000249989 UP000037069 UP000001070 UP000008792 UP000268350 UP000030765 UP000078200 UP000092460 UP000001819 UP000092445 UP000091820 UP000076408 UP000027135 UP000009192 UP000095300 UP000008237 UP000095301 UP000015103 UP000192223 UP000092553 UP000019118 UP000235965 UP000005203 UP000075903 UP000053825 UP000007266 UP000242457 UP000053097 UP000007755 UP000036403 UP000000311 UP000215335 UP000009046 UP000002358 UP000198287 UP000271974 UP000094527 UP000092444 UP000001554
UP000075885 UP000000673 UP000075884 UP000002320 UP000075902 UP000076407 UP000075882 UP000007062 UP000075840 UP000008820 UP000192221 UP000000304 UP000000803 UP000002282 UP000007801 UP000007798 UP000001292 UP000008711 UP000069940 UP000249989 UP000037069 UP000001070 UP000008792 UP000268350 UP000030765 UP000078200 UP000092460 UP000001819 UP000092445 UP000091820 UP000076408 UP000027135 UP000009192 UP000095300 UP000008237 UP000095301 UP000015103 UP000192223 UP000092553 UP000019118 UP000235965 UP000005203 UP000075903 UP000053825 UP000007266 UP000242457 UP000053097 UP000007755 UP000036403 UP000000311 UP000215335 UP000009046 UP000002358 UP000198287 UP000271974 UP000094527 UP000092444 UP000001554
PRIDE
Pfam
Interpro
IPR003959
ATPase_AAA_core
+ More
IPR027417 P-loop_NTPase
IPR003960 ATPase_AAA_CS
IPR041569 AAA_lid_3
IPR003593 AAA+_ATPase
IPR032501 Prot_ATP_ID_OB
IPR035244 26S_subunit_4
IPR005937 26S_Psome_P45-like
IPR016024 ARM-type_fold
IPR007015 DNA_pol_V/MYBBP1A
IPR011547 SLC26A/SulP_dom
IPR036513 STAS_dom_sf
IPR001902 SLC26A/SulP_fam
IPR036390 WH_DNA-bd_sf
IPR036388 WH-like_DNA-bd_sf
IPR007832 RNA_pol_Rpc34
IPR002645 STAS_dom
IPR035595 UDP_glycos_trans_CS
IPR002213 UDP_glucos_trans
IPR013083 Znf_RING/FYVE/PHD
IPR001841 Znf_RING
IPR027417 P-loop_NTPase
IPR003960 ATPase_AAA_CS
IPR041569 AAA_lid_3
IPR003593 AAA+_ATPase
IPR032501 Prot_ATP_ID_OB
IPR035244 26S_subunit_4
IPR005937 26S_Psome_P45-like
IPR016024 ARM-type_fold
IPR007015 DNA_pol_V/MYBBP1A
IPR011547 SLC26A/SulP_dom
IPR036513 STAS_dom_sf
IPR001902 SLC26A/SulP_fam
IPR036390 WH_DNA-bd_sf
IPR036388 WH-like_DNA-bd_sf
IPR007832 RNA_pol_Rpc34
IPR002645 STAS_dom
IPR035595 UDP_glycos_trans_CS
IPR002213 UDP_glucos_trans
IPR013083 Znf_RING/FYVE/PHD
IPR001841 Znf_RING
Gene 3D
ProteinModelPortal
A0A2A4JHF8
A0A3S2NVS4
A0A2H1W1T9
A0A194PG29
S4PC80
A0A182FSK9
+ More
I4DND5 A0A1J1HPP4 A0A182QB88 T1E2M9 A0A2M3Z2A7 A0A2M4A938 A0A2M4BNF7 A0A182PE42 E3WN61 A0A182N5B1 B0WWV3 A0A182TUL9 A0A182XE26 A0A182LCH1 A0T4G5 A0A182HJ96 Q1HQY1 A0A023ES93 A0A1W4UK15 B4QSD0 A0A1Q3FDQ6 A0A0B4LIJ0 B4PL52 B3M322 B4NBQ9 B4HG06 B3P8C1 P48601 A0A182G9P6 A0A1L8DT25 A0A0L0BMZ9 B4JRP6 B4M0C9 A0A3B0KCN1 A0A084WSL1 D3TMJ5 A0A1A9UQ60 A0A1B0BZ88 Q298N1 A0A1L8EHU8 A0A1B0A8W2 A0A1A9W6H4 U5EX60 A0A182XUX0 A0A067RGZ7 A0A336LG10 B4K736 A0A1I8QEI4 W8B0V2 E2BWH6 T1P826 A0A069DUE0 R4FNS2 A0A1W4X6M8 A0A0M4EK14 J3JXA9 A0A0K8UH06 A0A034W729 A0A2J7R280 R4WD80 A0A0B4J2L7 A0A182V0H7 A0A224XNL3 A0A0L7R1J4 V9IG44 A0A1B6HWI1 D6WTT2 A0A2A3EQB0 A0A0C9RQW5 A0A2R7VYI9 A0A026W6M8 A0A0A9WW95 A0A1B6MFA0 A0A1B6DDK7 A0A1Y1KB25 F4W4Q7 A0A0J7L139 E2ALJ9 A0A232ETH9 A0A0P4VME7 T1JN31 V5G8R3 E0W054 K7J1D7 A0A0P4WS62 A0A0P5GTX9 A0A2P6KDC2 A0A226ESC3 A0A3S1BJS8 A0A1D2MMR6 A0A1B0FHF8 A0A0K8TN88 A0A0B7BB55 C3XS01
I4DND5 A0A1J1HPP4 A0A182QB88 T1E2M9 A0A2M3Z2A7 A0A2M4A938 A0A2M4BNF7 A0A182PE42 E3WN61 A0A182N5B1 B0WWV3 A0A182TUL9 A0A182XE26 A0A182LCH1 A0T4G5 A0A182HJ96 Q1HQY1 A0A023ES93 A0A1W4UK15 B4QSD0 A0A1Q3FDQ6 A0A0B4LIJ0 B4PL52 B3M322 B4NBQ9 B4HG06 B3P8C1 P48601 A0A182G9P6 A0A1L8DT25 A0A0L0BMZ9 B4JRP6 B4M0C9 A0A3B0KCN1 A0A084WSL1 D3TMJ5 A0A1A9UQ60 A0A1B0BZ88 Q298N1 A0A1L8EHU8 A0A1B0A8W2 A0A1A9W6H4 U5EX60 A0A182XUX0 A0A067RGZ7 A0A336LG10 B4K736 A0A1I8QEI4 W8B0V2 E2BWH6 T1P826 A0A069DUE0 R4FNS2 A0A1W4X6M8 A0A0M4EK14 J3JXA9 A0A0K8UH06 A0A034W729 A0A2J7R280 R4WD80 A0A0B4J2L7 A0A182V0H7 A0A224XNL3 A0A0L7R1J4 V9IG44 A0A1B6HWI1 D6WTT2 A0A2A3EQB0 A0A0C9RQW5 A0A2R7VYI9 A0A026W6M8 A0A0A9WW95 A0A1B6MFA0 A0A1B6DDK7 A0A1Y1KB25 F4W4Q7 A0A0J7L139 E2ALJ9 A0A232ETH9 A0A0P4VME7 T1JN31 V5G8R3 E0W054 K7J1D7 A0A0P4WS62 A0A0P5GTX9 A0A2P6KDC2 A0A226ESC3 A0A3S1BJS8 A0A1D2MMR6 A0A1B0FHF8 A0A0K8TN88 A0A0B7BB55 C3XS01
PDB
6MSK
E-value=0,
Score=1887
Ontologies
KEGG
GO
GO:0036402
GO:0030163
GO:0005737
GO:0005524
GO:0008233
GO:0003677
GO:0005730
GO:0006355
GO:0008134
GO:0000502
GO:0006383
GO:0005666
GO:0016021
GO:0008271
GO:0017025
GO:0008540
GO:0043161
GO:0005838
GO:0008063
GO:0005654
GO:0005829
GO:0007623
GO:0016887
GO:0008568
GO:0016758
GO:0016787
GO:0005525
GO:0000166
GO:0008408
GO:0005112
GO:0015930
PANTHER
Topology
Subcellular location
Cytoplasm
Nucleus
Nucleus
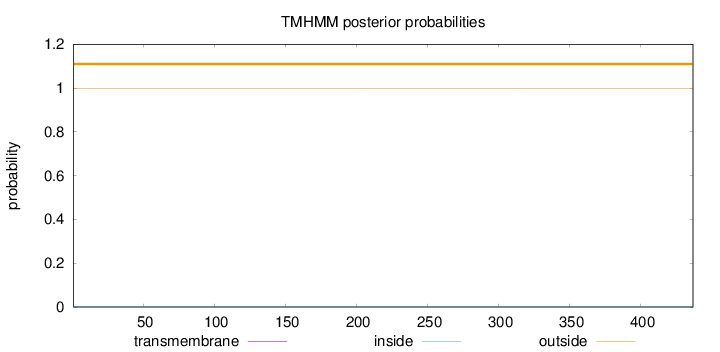
Length:
437
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00094
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00208
outside
1 - 437
Population Genetic Test Statistics
Pi
257.569066
Theta
221.666626
Tajima's D
0.632663
CLR
0
CSRT
0.550222488875556
Interpretation
Uncertain
