Pre Gene Modal
BGIBMGA004993
Annotation
putative_signal_recognition_particle_54_kDa_protein_[Bombyx_mori]
Full name
Signal recognition particle 54 kDa protein
Location in the cell
Cytoplasmic Reliability : 3.484
Sequence
CDS
ATGGTTTTAGCAGATTTAGGGCGTAAAATAACCACTGCTTTGCAGTCGTTGAGCAGGGCTACTATTATAAATGAAGAGGTATTGAATTCTATGCTGAAGCAGATATGTGCTGCATTATTGGAAGCAGATGTAAATATCCGCCTTGTAAAAAACTTAAGAGAAAATGTTCGCGCTGTCATTGATTTTGATGAAATGGCTGGTGGACTTAATAAGCGAAGGATGATTCAGTCTGCGGTTTTCAAAGAACTAGTCAAGCTTGTAGATCCCGGGGTTAAACCATACCAGCCCGTCAAAGGGAAGCCTAATGTAATAATGTTTGTGGGATTACAAGGATCTGGAAAGACCACAACATGTACAAAACTTGCATATCATTACCTACGTAAGAATTGGAAATCATGTTTGGTATGTGCTGACACATTCAGAGCTGGTGCTTATGATCAAGTCAAACAGAATTGTACTAAAGCCAGAATTCCGTTTTATGGAAGCTACACAGAGGTGGATCCAGTAGTAATAGCAACAACTGGAGTGGATATGTTCAAGAAGCAAGGTTTTGAAATGATTATTGTGGATACTAGTGGTCGTCATAAGCAAGAAGAGTCGCTTTTTGAAGAGATGTTGGCAGTTGCTACAGCTATTAAACCAGACAACATAATTTTTGTGATGGATGCAACAATTGGTCAAGCTTGTGAAGCTCAAGCCAGGGCTTTCAAAGACAAAGTTGATATTGGTTCTGTTATCATTACAAAGTTAGATGGACATGCAAAAGGAGGAGGTGCACTTTCGGCTGTTGCAGCTACACAAAGTCCTATCATATTCATTGGTACTGGAGAACATATTGATGACTTAGAGCCTTTCAAAACAAAACCGTTTATAAAGAAACTCCTTGGCATGGGAGACATTGAAGGTCTCTTAGATAAAGTTAATGAACTCAAACTCGATGACAATGATGAGCTACTAGAAAAACTTAAACATGGACAGTTTACTCTCAGAGCGATGTATGAACAATTTCAAAATATAATGAAGATGGGGCCTTTCTCACAAATAATGGGAATGATTCCTGGATTTTCACAAGATCTTATGTCAAAAGGAAGTGAACAAGAGTCAATGGCCAAATTAAAACTCCTTATGACAATTATGGACTCTATGAATGAAGGTGAGCTGGATAATAGAGACGGCGCCAAGTTGTTTTCTAAACAACCGACACGTATCACAAGAGTTGCACAGGGAGCAGGTGTCACAGAACGAGATGTTAAAGACTTAATTGCACAATACACAAAATTTGCGGCTGTCGTTAAAAAAATGGGTGGAATTAAAGGATTGTTCAAAGGAGGAGATATGGCTAAGAATGTTAATCAAACACAAATGGTTAAATTGAACCAACAAATGGCAAAGATGATGGATCCTCGCCTCCTACAACAAATGGGTGGTATGTCAGGTTTACACAATATGATGCGACAGCTACAACAGGGCTCAAGTGGAATGGGTGGGTTAATGGGTGGCAAATGA
Protein
MVLADLGRKITTALQSLSRATIINEEVLNSMLKQICAALLEADVNIRLVKNLRENVRAVIDFDEMAGGLNKRRMIQSAVFKELVKLVDPGVKPYQPVKGKPNVIMFVGLQGSGKTTTCTKLAYHYLRKNWKSCLVCADTFRAGAYDQVKQNCTKARIPFYGSYTEVDPVVIATTGVDMFKKQGFEMIIVDTSGRHKQEESLFEEMLAVATAIKPDNIIFVMDATIGQACEAQARAFKDKVDIGSVIITKLDGHAKGGGALSAVAATQSPIIFIGTGEHIDDLEPFKTKPFIKKLLGMGDIEGLLDKVNELKLDDNDELLEKLKHGQFTLRAMYEQFQNIMKMGPFSQIMGMIPGFSQDLMSKGSEQESMAKLKLLMTIMDSMNEGELDNRDGAKLFSKQPTRITRVAQGAGVTERDVKDLIAQYTKFAAVVKKMGGIKGLFKGGDMAKNVNQTQMVKLNQQMAKMMDPRLLQQMGGMSGLHNMMRQLQQGSSGMGGLMGGK
Summary
Description
Binds to the signal sequence of presecretory protein when they emerge from the ribosomes and transfers them to TRAM (translocating chain-associating membrane protein).
Subunit
Signal recognition particle consists of a 7S RNA molecule and at least six protein subunits.
Similarity
Belongs to the tRNA nucleotidyltransferase/poly(A) polymerase family.
Belongs to the GTP-binding SRP family. SRP54 subfamily.
Belongs to the peptidase S1 family.
Belongs to the GTP-binding SRP family. SRP54 subfamily.
Belongs to the peptidase S1 family.
Uniprot
H9J652
Q0N2R8
A0A212FKX0
A0A2A4JJ20
S4P9G7
A0A2H1W1X2
+ More
A0A194PFU1 A0A194RQE3 V5GTI2 A0A1Y1N6I0 A0A1L8DWB8 A0A2D1QUC4 D6WFU9 A0A336MTG6 A0A1B6G6U6 U5EWA7 A0A1B6HQ90 A0A232F6T2 A0A1B6KUP8 A0A084W7T8 A0A182H825 A0A067R011 A0A0P4VUR5 A0A0V0G4F3 A0A023F5T9 T1HX96 A0A182IN25 A0A2J7Q4U6 A0A2M3ZGU9 A0A2M4A3Y4 A0A2M4BKU1 W5JS33 A0A224XF31 A0A182MHU1 A0A182YEU3 A0A1B6DF04 A0A182PQA5 A0A182UVX1 A0A182VPP8 A0A182TEC6 A0A182XDP7 A0A182LNM8 Q7Q7N8 A0A182REC2 A0A182IDF4 A0A182NMK1 A0A182QN33 A0A2M4A4B3 A0A182FP74 A0A1Q3FCN1 B0WQI7 T1PH20 A0A0A9WER6 A0A1I8NLA4 A0A088AFN8 J3JTE3 E2BWT7 A0A026WIP8 A0A0J7NJR5 E2A7N3 E0VGY3 A0A151J0I4 F4WMJ0 A0A151I0Y6 A0A195EWA8 A0A158NAE8 A0A1L8ECS0 A0A0P5YU35 A0A0P5BCW9 A0A0N7ZT08 A0A0P6INB2 E9H4K5 A0A0P5V9L8 A0A195CJY6 A0A151X4X8 A0A0N7ZWJ4 A0A0L0C9J9 A0A0P5U7R3 A0A0K8TT08 A0A2A3ECY5 A0A1A9W7G5 A0A1B0G1Y1 A0A1A9XV31 A0A1A9VJ67 A0A1B0B988 A0A1B0AEU9 A0A3L8DDN6 A0A1W4US40 D3TNF4 B4N5H4 A0A0P5TT54 A0A1J1I3V1 A0A0P5BGR2 A0A1D2NGG5 B3M3R7 A0A3B0KFM8 W8BMN5 Q29FC0 B4H1J1 A0A2S2PWQ4
A0A194PFU1 A0A194RQE3 V5GTI2 A0A1Y1N6I0 A0A1L8DWB8 A0A2D1QUC4 D6WFU9 A0A336MTG6 A0A1B6G6U6 U5EWA7 A0A1B6HQ90 A0A232F6T2 A0A1B6KUP8 A0A084W7T8 A0A182H825 A0A067R011 A0A0P4VUR5 A0A0V0G4F3 A0A023F5T9 T1HX96 A0A182IN25 A0A2J7Q4U6 A0A2M3ZGU9 A0A2M4A3Y4 A0A2M4BKU1 W5JS33 A0A224XF31 A0A182MHU1 A0A182YEU3 A0A1B6DF04 A0A182PQA5 A0A182UVX1 A0A182VPP8 A0A182TEC6 A0A182XDP7 A0A182LNM8 Q7Q7N8 A0A182REC2 A0A182IDF4 A0A182NMK1 A0A182QN33 A0A2M4A4B3 A0A182FP74 A0A1Q3FCN1 B0WQI7 T1PH20 A0A0A9WER6 A0A1I8NLA4 A0A088AFN8 J3JTE3 E2BWT7 A0A026WIP8 A0A0J7NJR5 E2A7N3 E0VGY3 A0A151J0I4 F4WMJ0 A0A151I0Y6 A0A195EWA8 A0A158NAE8 A0A1L8ECS0 A0A0P5YU35 A0A0P5BCW9 A0A0N7ZT08 A0A0P6INB2 E9H4K5 A0A0P5V9L8 A0A195CJY6 A0A151X4X8 A0A0N7ZWJ4 A0A0L0C9J9 A0A0P5U7R3 A0A0K8TT08 A0A2A3ECY5 A0A1A9W7G5 A0A1B0G1Y1 A0A1A9XV31 A0A1A9VJ67 A0A1B0B988 A0A1B0AEU9 A0A3L8DDN6 A0A1W4US40 D3TNF4 B4N5H4 A0A0P5TT54 A0A1J1I3V1 A0A0P5BGR2 A0A1D2NGG5 B3M3R7 A0A3B0KFM8 W8BMN5 Q29FC0 B4H1J1 A0A2S2PWQ4
Pubmed
19121390
22118469
23622113
26354079
28004739
18362917
+ More
19820115 28648823 24438588 26483478 24845553 27129103 25474469 20920257 23761445 25244985 20966253 12364791 14747013 17210077 25315136 25401762 26823975 22516182 23537049 20798317 24508170 20566863 21719571 21347285 21292972 26108605 26369729 30249741 20353571 17994087 27289101 24495485 15632085
19820115 28648823 24438588 26483478 24845553 27129103 25474469 20920257 23761445 25244985 20966253 12364791 14747013 17210077 25315136 25401762 26823975 22516182 23537049 20798317 24508170 20566863 21719571 21347285 21292972 26108605 26369729 30249741 20353571 17994087 27289101 24495485 15632085
EMBL
BABH01030005
BABH01030006
BABH01030007
BABH01030008
DQ646413
ABH10803.1
+ More
AGBW02007949 OWR54391.1 NWSH01001392 PCG71410.1 GAIX01005491 JAA87069.1 ODYU01005792 SOQ47027.1 KQ459604 KPI92251.1 KQ460045 KPJ18246.1 GALX01003534 JAB64932.1 GEZM01013898 JAV92265.1 GFDF01003539 JAV10545.1 KY285043 ATP16146.1 KQ971328 EEZ99794.1 UFQS01002172 UFQT01002172 SSX13407.1 SSX32841.1 GECZ01011716 GECZ01003614 JAS58053.1 JAS66155.1 GANO01001507 JAB58364.1 GECU01030872 JAS76834.1 NNAY01000855 OXU26190.1 GEBQ01024856 JAT15121.1 ATLV01021297 KE525316 KFB46282.1 JXUM01117524 KQ566093 KXJ70403.1 KK852804 KDR16155.1 GDKW01000821 JAI55774.1 GECL01003556 JAP02568.1 GBBI01002169 JAC16543.1 ACPB03001298 NEVH01018377 PNF23601.1 GGFM01006980 MBW27731.1 GGFK01002203 MBW35524.1 GGFJ01004546 MBW53687.1 ADMH02000585 ETN65735.1 GFTR01006802 JAW09624.1 AXCM01000109 GEDC01013014 JAS24284.1 AAAB01008952 EAA10561.2 APCN01005971 AXCN02001594 GGFK01002274 MBW35595.1 GFDL01009782 JAV25263.1 DS232041 EDS32862.1 KA647188 AFP61817.1 GBHO01040244 GBHO01040242 GDHC01014169 JAG03360.1 JAG03362.1 JAQ04460.1 APGK01034495 BT126499 KB740914 KB632005 AEE61463.1 ENN78340.1 ERL87923.1 GL451188 EFN79801.1 KK107231 EZA54974.1 LBMM01004181 KMQ92725.1 GL437365 EFN70554.1 AAZO01002262 DS235154 EEB12639.1 KQ980629 KYN14994.1 GL888217 EGI64601.1 KQ976600 KYM79474.1 KQ981954 KYN32174.1 ADTU01010270 ADTU01010271 GFDG01002367 JAV16432.1 GDIP01105337 GDIP01053410 LRGB01000146 JAM50305.1 KZS20586.1 GDIP01191592 JAJ31810.1 GDIP01215473 JAJ07929.1 GDIQ01001543 JAN93194.1 GL732592 EFX73155.1 GDIP01102541 JAM01174.1 KQ977642 KYN01041.1 KQ982519 KYQ55485.1 GDIP01205585 JAJ17817.1 JRES01000692 KNC29113.1 GDIP01116833 JAL86881.1 GDAI01000325 JAI17278.1 KZ288301 PBC28881.1 CCAG010004398 JXJN01010304 QOIP01000009 RLU18600.1 EZ422956 ADD19232.1 CH964101 EDW79613.1 GDIP01127342 JAL76372.1 CVRI01000040 CRK94973.1 GDIP01189075 JAJ34327.1 LJIJ01000047 ODN04339.1 CH902618 EDV40360.1 OUUW01000012 SPP87160.1 GAMC01004200 JAC02356.1 CH379067 EAL31335.1 CH479202 EDW30168.1 GGMS01000587 MBY69790.1
AGBW02007949 OWR54391.1 NWSH01001392 PCG71410.1 GAIX01005491 JAA87069.1 ODYU01005792 SOQ47027.1 KQ459604 KPI92251.1 KQ460045 KPJ18246.1 GALX01003534 JAB64932.1 GEZM01013898 JAV92265.1 GFDF01003539 JAV10545.1 KY285043 ATP16146.1 KQ971328 EEZ99794.1 UFQS01002172 UFQT01002172 SSX13407.1 SSX32841.1 GECZ01011716 GECZ01003614 JAS58053.1 JAS66155.1 GANO01001507 JAB58364.1 GECU01030872 JAS76834.1 NNAY01000855 OXU26190.1 GEBQ01024856 JAT15121.1 ATLV01021297 KE525316 KFB46282.1 JXUM01117524 KQ566093 KXJ70403.1 KK852804 KDR16155.1 GDKW01000821 JAI55774.1 GECL01003556 JAP02568.1 GBBI01002169 JAC16543.1 ACPB03001298 NEVH01018377 PNF23601.1 GGFM01006980 MBW27731.1 GGFK01002203 MBW35524.1 GGFJ01004546 MBW53687.1 ADMH02000585 ETN65735.1 GFTR01006802 JAW09624.1 AXCM01000109 GEDC01013014 JAS24284.1 AAAB01008952 EAA10561.2 APCN01005971 AXCN02001594 GGFK01002274 MBW35595.1 GFDL01009782 JAV25263.1 DS232041 EDS32862.1 KA647188 AFP61817.1 GBHO01040244 GBHO01040242 GDHC01014169 JAG03360.1 JAG03362.1 JAQ04460.1 APGK01034495 BT126499 KB740914 KB632005 AEE61463.1 ENN78340.1 ERL87923.1 GL451188 EFN79801.1 KK107231 EZA54974.1 LBMM01004181 KMQ92725.1 GL437365 EFN70554.1 AAZO01002262 DS235154 EEB12639.1 KQ980629 KYN14994.1 GL888217 EGI64601.1 KQ976600 KYM79474.1 KQ981954 KYN32174.1 ADTU01010270 ADTU01010271 GFDG01002367 JAV16432.1 GDIP01105337 GDIP01053410 LRGB01000146 JAM50305.1 KZS20586.1 GDIP01191592 JAJ31810.1 GDIP01215473 JAJ07929.1 GDIQ01001543 JAN93194.1 GL732592 EFX73155.1 GDIP01102541 JAM01174.1 KQ977642 KYN01041.1 KQ982519 KYQ55485.1 GDIP01205585 JAJ17817.1 JRES01000692 KNC29113.1 GDIP01116833 JAL86881.1 GDAI01000325 JAI17278.1 KZ288301 PBC28881.1 CCAG010004398 JXJN01010304 QOIP01000009 RLU18600.1 EZ422956 ADD19232.1 CH964101 EDW79613.1 GDIP01127342 JAL76372.1 CVRI01000040 CRK94973.1 GDIP01189075 JAJ34327.1 LJIJ01000047 ODN04339.1 CH902618 EDV40360.1 OUUW01000012 SPP87160.1 GAMC01004200 JAC02356.1 CH379067 EAL31335.1 CH479202 EDW30168.1 GGMS01000587 MBY69790.1
Proteomes
UP000005204
UP000007151
UP000218220
UP000053268
UP000053240
UP000007266
+ More
UP000215335 UP000030765 UP000069940 UP000249989 UP000027135 UP000015103 UP000075880 UP000235965 UP000000673 UP000075883 UP000076408 UP000075885 UP000075903 UP000075920 UP000075902 UP000076407 UP000075882 UP000007062 UP000075900 UP000075840 UP000075884 UP000075886 UP000069272 UP000002320 UP000095301 UP000095300 UP000005203 UP000019118 UP000030742 UP000008237 UP000053097 UP000036403 UP000000311 UP000009046 UP000078492 UP000007755 UP000078540 UP000078541 UP000005205 UP000076858 UP000000305 UP000078542 UP000075809 UP000037069 UP000242457 UP000091820 UP000092444 UP000092443 UP000078200 UP000092460 UP000092445 UP000279307 UP000192221 UP000007798 UP000183832 UP000094527 UP000007801 UP000268350 UP000001819 UP000008744
UP000215335 UP000030765 UP000069940 UP000249989 UP000027135 UP000015103 UP000075880 UP000235965 UP000000673 UP000075883 UP000076408 UP000075885 UP000075903 UP000075920 UP000075902 UP000076407 UP000075882 UP000007062 UP000075900 UP000075840 UP000075884 UP000075886 UP000069272 UP000002320 UP000095301 UP000095300 UP000005203 UP000019118 UP000030742 UP000008237 UP000053097 UP000036403 UP000000311 UP000009046 UP000078492 UP000007755 UP000078540 UP000078541 UP000005205 UP000076858 UP000000305 UP000078542 UP000075809 UP000037069 UP000242457 UP000091820 UP000092444 UP000092443 UP000078200 UP000092460 UP000092445 UP000279307 UP000192221 UP000007798 UP000183832 UP000094527 UP000007801 UP000268350 UP000001819 UP000008744
PRIDE
Pfam
Interpro
IPR036225
SRP/SRP_N
+ More
IPR036428 PCD_sf
IPR022941 SRP54
IPR004125 Signal_recog_particle_SRP54_M
IPR036891 Signal_recog_part_SRP54_M_sf
IPR003593 AAA+_ATPase
IPR000897 SRP54_GTPase_dom
IPR001533 Pterin_deHydtase
IPR042101 SRP54_N_sf
IPR002646 PolA_pol_head_dom
IPR006325 SRP54_euk
IPR013822 Signal_recog_particl_SRP54_hlx
IPR027417 P-loop_NTPase
IPR013087 Znf_C2H2_type
IPR036236 Znf_C2H2_sf
IPR018114 TRYPSIN_HIS
IPR001254 Trypsin_dom
IPR009003 Peptidase_S1_PA
IPR001314 Peptidase_S1A
IPR022700 CLIP
IPR036428 PCD_sf
IPR022941 SRP54
IPR004125 Signal_recog_particle_SRP54_M
IPR036891 Signal_recog_part_SRP54_M_sf
IPR003593 AAA+_ATPase
IPR000897 SRP54_GTPase_dom
IPR001533 Pterin_deHydtase
IPR042101 SRP54_N_sf
IPR002646 PolA_pol_head_dom
IPR006325 SRP54_euk
IPR013822 Signal_recog_particl_SRP54_hlx
IPR027417 P-loop_NTPase
IPR013087 Znf_C2H2_type
IPR036236 Znf_C2H2_sf
IPR018114 TRYPSIN_HIS
IPR001254 Trypsin_dom
IPR009003 Peptidase_S1_PA
IPR001314 Peptidase_S1A
IPR022700 CLIP
SUPFAM
Gene 3D
CDD
ProteinModelPortal
H9J652
Q0N2R8
A0A212FKX0
A0A2A4JJ20
S4P9G7
A0A2H1W1X2
+ More
A0A194PFU1 A0A194RQE3 V5GTI2 A0A1Y1N6I0 A0A1L8DWB8 A0A2D1QUC4 D6WFU9 A0A336MTG6 A0A1B6G6U6 U5EWA7 A0A1B6HQ90 A0A232F6T2 A0A1B6KUP8 A0A084W7T8 A0A182H825 A0A067R011 A0A0P4VUR5 A0A0V0G4F3 A0A023F5T9 T1HX96 A0A182IN25 A0A2J7Q4U6 A0A2M3ZGU9 A0A2M4A3Y4 A0A2M4BKU1 W5JS33 A0A224XF31 A0A182MHU1 A0A182YEU3 A0A1B6DF04 A0A182PQA5 A0A182UVX1 A0A182VPP8 A0A182TEC6 A0A182XDP7 A0A182LNM8 Q7Q7N8 A0A182REC2 A0A182IDF4 A0A182NMK1 A0A182QN33 A0A2M4A4B3 A0A182FP74 A0A1Q3FCN1 B0WQI7 T1PH20 A0A0A9WER6 A0A1I8NLA4 A0A088AFN8 J3JTE3 E2BWT7 A0A026WIP8 A0A0J7NJR5 E2A7N3 E0VGY3 A0A151J0I4 F4WMJ0 A0A151I0Y6 A0A195EWA8 A0A158NAE8 A0A1L8ECS0 A0A0P5YU35 A0A0P5BCW9 A0A0N7ZT08 A0A0P6INB2 E9H4K5 A0A0P5V9L8 A0A195CJY6 A0A151X4X8 A0A0N7ZWJ4 A0A0L0C9J9 A0A0P5U7R3 A0A0K8TT08 A0A2A3ECY5 A0A1A9W7G5 A0A1B0G1Y1 A0A1A9XV31 A0A1A9VJ67 A0A1B0B988 A0A1B0AEU9 A0A3L8DDN6 A0A1W4US40 D3TNF4 B4N5H4 A0A0P5TT54 A0A1J1I3V1 A0A0P5BGR2 A0A1D2NGG5 B3M3R7 A0A3B0KFM8 W8BMN5 Q29FC0 B4H1J1 A0A2S2PWQ4
A0A194PFU1 A0A194RQE3 V5GTI2 A0A1Y1N6I0 A0A1L8DWB8 A0A2D1QUC4 D6WFU9 A0A336MTG6 A0A1B6G6U6 U5EWA7 A0A1B6HQ90 A0A232F6T2 A0A1B6KUP8 A0A084W7T8 A0A182H825 A0A067R011 A0A0P4VUR5 A0A0V0G4F3 A0A023F5T9 T1HX96 A0A182IN25 A0A2J7Q4U6 A0A2M3ZGU9 A0A2M4A3Y4 A0A2M4BKU1 W5JS33 A0A224XF31 A0A182MHU1 A0A182YEU3 A0A1B6DF04 A0A182PQA5 A0A182UVX1 A0A182VPP8 A0A182TEC6 A0A182XDP7 A0A182LNM8 Q7Q7N8 A0A182REC2 A0A182IDF4 A0A182NMK1 A0A182QN33 A0A2M4A4B3 A0A182FP74 A0A1Q3FCN1 B0WQI7 T1PH20 A0A0A9WER6 A0A1I8NLA4 A0A088AFN8 J3JTE3 E2BWT7 A0A026WIP8 A0A0J7NJR5 E2A7N3 E0VGY3 A0A151J0I4 F4WMJ0 A0A151I0Y6 A0A195EWA8 A0A158NAE8 A0A1L8ECS0 A0A0P5YU35 A0A0P5BCW9 A0A0N7ZT08 A0A0P6INB2 E9H4K5 A0A0P5V9L8 A0A195CJY6 A0A151X4X8 A0A0N7ZWJ4 A0A0L0C9J9 A0A0P5U7R3 A0A0K8TT08 A0A2A3ECY5 A0A1A9W7G5 A0A1B0G1Y1 A0A1A9XV31 A0A1A9VJ67 A0A1B0B988 A0A1B0AEU9 A0A3L8DDN6 A0A1W4US40 D3TNF4 B4N5H4 A0A0P5TT54 A0A1J1I3V1 A0A0P5BGR2 A0A1D2NGG5 B3M3R7 A0A3B0KFM8 W8BMN5 Q29FC0 B4H1J1 A0A2S2PWQ4
PDB
6FRK
E-value=0,
Score=1938
Ontologies
GO
PANTHER
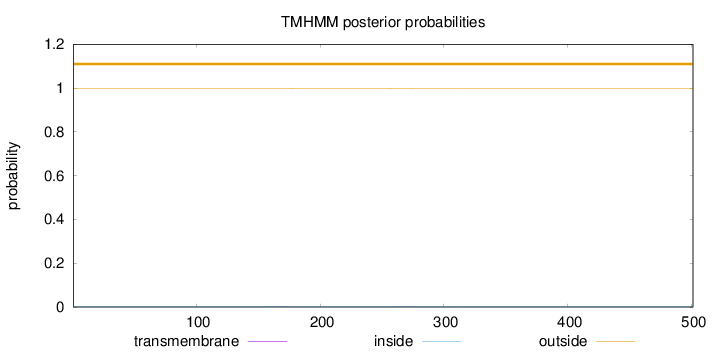
Topology
Subcellular location
Cytoplasm
Length:
501
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0273399999999999
Exp number, first 60 AAs:
0.00023
Total prob of N-in:
0.00278
outside
1 - 501
Population Genetic Test Statistics
Pi
14.990922
Theta
18.742286
Tajima's D
-0.614738
CLR
22.572305
CSRT
0.221638918054097
Interpretation
Uncertain
