Gene
KWMTBOMO15120 Validated by peptides from experiments
Annotation
PREDICTED:_CCA_tRNA_nucleotidyltransferase_1?_mitochondrial_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 3.29
Sequence
CDS
ATGGACTTAAATTTAAAGTGTCGGGAAAATCCTATTGTTATTAAAGTGGATTCGCCAGAGTTTAAAAGTATATTTACAGAAGAAGTTATAGACCTAAAAAAAATTTTTGACAAATATAAACATGAAATTAGAATAGCTGGTGGTGCTGTAAGAGATCTTTTAATGGGACTTACACCAAAAGATCTTGATTTTGCCACAACAGCAACTCCTGAAGAAATGAAAGAAATGTTCACTGCGGAAGAAGTAAGAATGATAAACGTTAATGGAGAGAGGCATGGAACTATCACTGCTCGAATTAACGACAAGGAGAATTTTGAGGTAACCACCTTAAGGATTGATATGGTTACCGATGGACGTTTTGCAGATGTTCAATTCACTAAAGATTGGAAACTTGATGCTAACAGAAGAGACCTTACTATAAATTCTATGTTTTTAGGGTTTGATGGATCTGTTTACGATTACTTTTATGGATATGAAGATTTACAAAAGAGAAGAGTTGCATTTGTTGGAGATCCTGATGTTAGAGTCAAAGAAGATTACTTGAGAATTATGCGTTATTTCCGATTTTATGGAAGAATAGCACTAAAACCAGACAATCATGAGGAAGAGACACTAAGAGTACTTAAAAATAATGTTCATGGCTTGGAAAATATATCAGGTGAAAGAATATGGGGTGAACTTAAAAAAATTTTGCAAGGAAACTTTGCTGGAAATTTGTTGAAGACTATGCTCAATGTAGGTGTAGGAGAGTACATGGGTTTCCCATTATCTCCTAATGTGCAAGAACTGGATATGTTATTAAAAAGAGCCCATCATCTTAATTTGAGTCCACTCAGTTATCTTGCTGCTCTAGTTAAAGATTTAGACGAAGTCACAGTCTTGCATAGCAGAATGAAATTCTCGAAATTGGAAAGAGATTTGCTTTACTTTCTGGTAGAACATAGAGATGACAAAGTTTCTGATAGACCCATGAAACCCTATGAATCATTAATATTAAATACGTCAATGAAGCAAAAAGTAGCCATAGAATATGTGCGAGAAGTTTTAAAGTATAGAGGAGATGAAGAACTCTTAAAAAGACTTGACAACTGGGAAATCCCTCGTTTCCCTGTCTCTGGAAAAACCCTTAAAGAATGTGGAGTACCAGTTGGGAAAATAAAAAGACTTAATGTCTTATTTACCTAA
Protein
MDLNLKCRENPIVIKVDSPEFKSIFTEEVIDLKKIFDKYKHEIRIAGGAVRDLLMGLTPKDLDFATTATPEEMKEMFTAEEVRMINVNGERHGTITARINDKENFEVTTLRIDMVTDGRFADVQFTKDWKLDANRRDLTINSMFLGFDGSVYDYFYGYEDLQKRRVAFVGDPDVRVKEDYLRIMRYFRFYGRIALKPDNHEEETLRVLKNNVHGLENISGERIWGELKKILQGNFAGNLLKTMLNVGVGEYMGFPLSPNVQELDMLLKRAHHLNLSPLSYLAALVKDLDEVTVLHSRMKFSKLERDLLYFLVEHRDDKVSDRPMKPYESLILNTSMKQKVAIEYVREVLKYRGDEELLKRLDNWEIPRFPVSGKTLKECGVPVGKIKRLNVLFT
Summary
Similarity
Belongs to the tRNA nucleotidyltransferase/poly(A) polymerase family.
Uniprot
A0A2H1W247
A0A194RK90
A0A194PHQ2
A0A212FKY0
A0A0L7KSR2
A0A1Y1KLT4
+ More
A0A067QMS9 A0A2J7R6Q4 A0A2J7R6R1 A0A182PCD2 D6WPQ0 A0A1Q3F6J0 A0A0P6IUW3 B0X1I4 A0A182RLE7 N6SSP0 U4UQC7 A0A182MBI9 A0A0C9QHY9 A0A182W0W7 A0A182HSK4 A0A182XAR2 Q7QCF2 A0A182QZM7 A0A0J7KG76 E2B0H7 A0A182JGI0 A0A182VN68 A0A182TXN6 A0A2R5LNQ0 A0A0K8TSF9 A0A131YKX4 T1HLF6 A0A182NRH5 R4G8X9 A0A224Z2H8 A0A336M097 L7M851 U5EXI2 A0A023FX51 A0A182TAR1 A0A131Y076 A0A026X2T0 A0A182FT11 B7Q4B3 A0A1A9X1T7 A0A1W4WS05 W5JUZ3 A0A182Y6I7 A0A195CYG6 A0A195EHU9 A0A069DU53 A0A1L8DRX1 A0A1E1XCL4 A0A0V0G9C8 A0A2R7X2A3 D3TPV7 F4W8F9 A0A023F915 A0A1A9Z6X6 A0A0L0CFA3 A0A1I8PR30 A0A1B0BXD2 E2BW48 A0A158NK65 A0A1Z5KV19 A0A1A9YBV0 A0A1A9US60 A0A151X6P4 A0A232ELV3 A0A0K8UI83 A0A1B6KQW0 A0A0K8V0J5 A0A1I8NDD8 A0A023GKF0 G3MLP9 A0A293MQG1 W8BNP5 E9H6E9 A0A0A9Y0H3 B4KCB6 A0A034W977 K7J2Z4 A0A0M3QX99 A0A0P5PRQ2 A0A0P5JIZ7 A0A0A1WVK8 A0A0P5QU46 A0A0P5CF70 A0A0P5PAC4 A0A0P5M6M5 A0A0P5RU68 A0A0P5QZG6 A0A0P5VF14 W4YX98 B3LWE9 B4LZ70 A0A1B6DJ94 B4JUH0 A0A0L7RHD8
A0A067QMS9 A0A2J7R6Q4 A0A2J7R6R1 A0A182PCD2 D6WPQ0 A0A1Q3F6J0 A0A0P6IUW3 B0X1I4 A0A182RLE7 N6SSP0 U4UQC7 A0A182MBI9 A0A0C9QHY9 A0A182W0W7 A0A182HSK4 A0A182XAR2 Q7QCF2 A0A182QZM7 A0A0J7KG76 E2B0H7 A0A182JGI0 A0A182VN68 A0A182TXN6 A0A2R5LNQ0 A0A0K8TSF9 A0A131YKX4 T1HLF6 A0A182NRH5 R4G8X9 A0A224Z2H8 A0A336M097 L7M851 U5EXI2 A0A023FX51 A0A182TAR1 A0A131Y076 A0A026X2T0 A0A182FT11 B7Q4B3 A0A1A9X1T7 A0A1W4WS05 W5JUZ3 A0A182Y6I7 A0A195CYG6 A0A195EHU9 A0A069DU53 A0A1L8DRX1 A0A1E1XCL4 A0A0V0G9C8 A0A2R7X2A3 D3TPV7 F4W8F9 A0A023F915 A0A1A9Z6X6 A0A0L0CFA3 A0A1I8PR30 A0A1B0BXD2 E2BW48 A0A158NK65 A0A1Z5KV19 A0A1A9YBV0 A0A1A9US60 A0A151X6P4 A0A232ELV3 A0A0K8UI83 A0A1B6KQW0 A0A0K8V0J5 A0A1I8NDD8 A0A023GKF0 G3MLP9 A0A293MQG1 W8BNP5 E9H6E9 A0A0A9Y0H3 B4KCB6 A0A034W977 K7J2Z4 A0A0M3QX99 A0A0P5PRQ2 A0A0P5JIZ7 A0A0A1WVK8 A0A0P5QU46 A0A0P5CF70 A0A0P5PAC4 A0A0P5M6M5 A0A0P5RU68 A0A0P5QZG6 A0A0P5VF14 W4YX98 B3LWE9 B4LZ70 A0A1B6DJ94 B4JUH0 A0A0L7RHD8
Pubmed
26354079
22118469
26227816
28004739
24845553
18362917
+ More
19820115 26999592 23537049 12364791 14747013 17210077 20798317 26369729 26830274 28797301 25576852 24508170 30249741 20920257 23761445 25244985 26334808 28503490 20353571 21719571 25474469 26108605 21347285 28528879 28648823 25315136 22216098 24495485 21292972 25401762 26823975 17994087 25348373 20075255 25830018
19820115 26999592 23537049 12364791 14747013 17210077 20798317 26369729 26830274 28797301 25576852 24508170 30249741 20920257 23761445 25244985 26334808 28503490 20353571 21719571 25474469 26108605 21347285 28528879 28648823 25315136 22216098 24495485 21292972 25401762 26823975 17994087 25348373 20075255 25830018
EMBL
ODYU01005792
SOQ47026.1
KQ460045
KPJ18248.1
KQ459604
KPI92249.1
+ More
AGBW02007949 OWR54392.1 JTDY01006327 KOB66101.1 GEZM01080226 JAV62284.1 KK853153 KDR10588.1 NEVH01006756 PNF36518.1 PNF36519.1 KQ971354 EFA06191.2 GFDL01011873 JAV23172.1 GDUN01000991 JAN94928.1 DS232260 EDS38633.1 APGK01058333 KB741288 ENN70684.1 KB632322 ERL92326.1 AXCM01000146 GBYB01014283 JAG84050.1 APCN01000281 AAAB01008859 EAA08067.5 AXCN02001459 LBMM01007938 KMQ89282.1 GL444608 EFN60819.1 GGLE01006801 MBY10927.1 GDAI01000530 JAI17073.1 GEDV01009382 JAP79175.1 ACPB03010982 GAHY01000192 JAA77318.1 GFPF01012871 MAA24017.1 UFQT01000380 SSX23746.1 GACK01004829 JAA60205.1 GANO01002527 JAB57344.1 GBBL01001223 JAC26097.1 GEFM01003921 JAP71875.1 KK107021 QOIP01000011 EZA62403.1 RLU16618.1 ABJB010408845 DS854427 EEC13685.1 ADMH02000418 ETN66594.1 KQ977110 KYN05698.1 KQ978881 KYN27828.1 GBGD01001593 JAC87296.1 GFDF01004882 JAV09202.1 GFAC01002206 JAT96982.1 GECL01001436 JAP04688.1 KK856508 PTY25922.1 CCAG010012181 EZ423459 ADD19735.1 GL887908 EGI69450.1 GBBI01001253 JAC17459.1 JRES01000562 KNC30184.1 JXJN01022181 GL451091 EFN80080.1 ADTU01018717 GFJQ02008018 JAV98951.1 KQ982472 KYQ56083.1 NNAY01003493 OXU19335.1 GDHF01025920 JAI26394.1 GEBQ01026149 JAT13828.1 GDHF01019943 JAI32371.1 GBBM01001057 JAC34361.1 JO842800 AEO34417.1 GFWV01023170 MAA47897.1 GAMC01011534 JAB95021.1 GL732597 EFX72643.1 GBHO01017032 GBRD01007414 GDHC01006352 JAG26572.1 JAG58407.1 JAQ12277.1 CH933806 EDW13725.2 GAKP01008619 GAKP01008617 JAC50333.1 CP012526 ALC45499.1 GDIQ01124279 JAL27447.1 GDIQ01202261 JAK49464.1 GBXI01011732 JAD02560.1 GDIQ01115888 JAL35838.1 GDIP01187187 LRGB01003375 JAJ36215.1 KZS02967.1 GDIQ01131392 JAL20334.1 GDIQ01160020 JAK91705.1 GDIQ01101543 JAL50183.1 GDIQ01113429 JAL38297.1 GDIP01102693 JAM01022.1 AAGJ04068193 CH902617 EDV43782.1 CH940650 EDW67077.2 GEDC01029396 GEDC01027642 GEDC01011527 GEDC01002698 JAS07902.1 JAS09656.1 JAS25771.1 JAS34600.1 CH916374 EDV91140.1 KQ414592 KOC70218.1
AGBW02007949 OWR54392.1 JTDY01006327 KOB66101.1 GEZM01080226 JAV62284.1 KK853153 KDR10588.1 NEVH01006756 PNF36518.1 PNF36519.1 KQ971354 EFA06191.2 GFDL01011873 JAV23172.1 GDUN01000991 JAN94928.1 DS232260 EDS38633.1 APGK01058333 KB741288 ENN70684.1 KB632322 ERL92326.1 AXCM01000146 GBYB01014283 JAG84050.1 APCN01000281 AAAB01008859 EAA08067.5 AXCN02001459 LBMM01007938 KMQ89282.1 GL444608 EFN60819.1 GGLE01006801 MBY10927.1 GDAI01000530 JAI17073.1 GEDV01009382 JAP79175.1 ACPB03010982 GAHY01000192 JAA77318.1 GFPF01012871 MAA24017.1 UFQT01000380 SSX23746.1 GACK01004829 JAA60205.1 GANO01002527 JAB57344.1 GBBL01001223 JAC26097.1 GEFM01003921 JAP71875.1 KK107021 QOIP01000011 EZA62403.1 RLU16618.1 ABJB010408845 DS854427 EEC13685.1 ADMH02000418 ETN66594.1 KQ977110 KYN05698.1 KQ978881 KYN27828.1 GBGD01001593 JAC87296.1 GFDF01004882 JAV09202.1 GFAC01002206 JAT96982.1 GECL01001436 JAP04688.1 KK856508 PTY25922.1 CCAG010012181 EZ423459 ADD19735.1 GL887908 EGI69450.1 GBBI01001253 JAC17459.1 JRES01000562 KNC30184.1 JXJN01022181 GL451091 EFN80080.1 ADTU01018717 GFJQ02008018 JAV98951.1 KQ982472 KYQ56083.1 NNAY01003493 OXU19335.1 GDHF01025920 JAI26394.1 GEBQ01026149 JAT13828.1 GDHF01019943 JAI32371.1 GBBM01001057 JAC34361.1 JO842800 AEO34417.1 GFWV01023170 MAA47897.1 GAMC01011534 JAB95021.1 GL732597 EFX72643.1 GBHO01017032 GBRD01007414 GDHC01006352 JAG26572.1 JAG58407.1 JAQ12277.1 CH933806 EDW13725.2 GAKP01008619 GAKP01008617 JAC50333.1 CP012526 ALC45499.1 GDIQ01124279 JAL27447.1 GDIQ01202261 JAK49464.1 GBXI01011732 JAD02560.1 GDIQ01115888 JAL35838.1 GDIP01187187 LRGB01003375 JAJ36215.1 KZS02967.1 GDIQ01131392 JAL20334.1 GDIQ01160020 JAK91705.1 GDIQ01101543 JAL50183.1 GDIQ01113429 JAL38297.1 GDIP01102693 JAM01022.1 AAGJ04068193 CH902617 EDV43782.1 CH940650 EDW67077.2 GEDC01029396 GEDC01027642 GEDC01011527 GEDC01002698 JAS07902.1 JAS09656.1 JAS25771.1 JAS34600.1 CH916374 EDV91140.1 KQ414592 KOC70218.1
Proteomes
UP000053240
UP000053268
UP000007151
UP000037510
UP000027135
UP000235965
+ More
UP000075885 UP000007266 UP000002320 UP000075900 UP000019118 UP000030742 UP000075883 UP000075920 UP000075840 UP000076407 UP000007062 UP000075886 UP000036403 UP000000311 UP000075880 UP000075903 UP000075902 UP000015103 UP000075884 UP000075901 UP000053097 UP000279307 UP000069272 UP000001555 UP000091820 UP000192223 UP000000673 UP000076408 UP000078542 UP000078492 UP000092444 UP000007755 UP000092445 UP000037069 UP000095300 UP000092460 UP000008237 UP000005205 UP000092443 UP000078200 UP000075809 UP000215335 UP000095301 UP000000305 UP000009192 UP000002358 UP000092553 UP000076858 UP000007110 UP000007801 UP000008792 UP000001070 UP000053825
UP000075885 UP000007266 UP000002320 UP000075900 UP000019118 UP000030742 UP000075883 UP000075920 UP000075840 UP000076407 UP000007062 UP000075886 UP000036403 UP000000311 UP000075880 UP000075903 UP000075902 UP000015103 UP000075884 UP000075901 UP000053097 UP000279307 UP000069272 UP000001555 UP000091820 UP000192223 UP000000673 UP000076408 UP000078542 UP000078492 UP000092444 UP000007755 UP000092445 UP000037069 UP000095300 UP000092460 UP000008237 UP000005205 UP000092443 UP000078200 UP000075809 UP000215335 UP000095301 UP000000305 UP000009192 UP000002358 UP000092553 UP000076858 UP000007110 UP000007801 UP000008792 UP000001070 UP000053825
Pfam
Interpro
Gene 3D
ProteinModelPortal
A0A2H1W247
A0A194RK90
A0A194PHQ2
A0A212FKY0
A0A0L7KSR2
A0A1Y1KLT4
+ More
A0A067QMS9 A0A2J7R6Q4 A0A2J7R6R1 A0A182PCD2 D6WPQ0 A0A1Q3F6J0 A0A0P6IUW3 B0X1I4 A0A182RLE7 N6SSP0 U4UQC7 A0A182MBI9 A0A0C9QHY9 A0A182W0W7 A0A182HSK4 A0A182XAR2 Q7QCF2 A0A182QZM7 A0A0J7KG76 E2B0H7 A0A182JGI0 A0A182VN68 A0A182TXN6 A0A2R5LNQ0 A0A0K8TSF9 A0A131YKX4 T1HLF6 A0A182NRH5 R4G8X9 A0A224Z2H8 A0A336M097 L7M851 U5EXI2 A0A023FX51 A0A182TAR1 A0A131Y076 A0A026X2T0 A0A182FT11 B7Q4B3 A0A1A9X1T7 A0A1W4WS05 W5JUZ3 A0A182Y6I7 A0A195CYG6 A0A195EHU9 A0A069DU53 A0A1L8DRX1 A0A1E1XCL4 A0A0V0G9C8 A0A2R7X2A3 D3TPV7 F4W8F9 A0A023F915 A0A1A9Z6X6 A0A0L0CFA3 A0A1I8PR30 A0A1B0BXD2 E2BW48 A0A158NK65 A0A1Z5KV19 A0A1A9YBV0 A0A1A9US60 A0A151X6P4 A0A232ELV3 A0A0K8UI83 A0A1B6KQW0 A0A0K8V0J5 A0A1I8NDD8 A0A023GKF0 G3MLP9 A0A293MQG1 W8BNP5 E9H6E9 A0A0A9Y0H3 B4KCB6 A0A034W977 K7J2Z4 A0A0M3QX99 A0A0P5PRQ2 A0A0P5JIZ7 A0A0A1WVK8 A0A0P5QU46 A0A0P5CF70 A0A0P5PAC4 A0A0P5M6M5 A0A0P5RU68 A0A0P5QZG6 A0A0P5VF14 W4YX98 B3LWE9 B4LZ70 A0A1B6DJ94 B4JUH0 A0A0L7RHD8
A0A067QMS9 A0A2J7R6Q4 A0A2J7R6R1 A0A182PCD2 D6WPQ0 A0A1Q3F6J0 A0A0P6IUW3 B0X1I4 A0A182RLE7 N6SSP0 U4UQC7 A0A182MBI9 A0A0C9QHY9 A0A182W0W7 A0A182HSK4 A0A182XAR2 Q7QCF2 A0A182QZM7 A0A0J7KG76 E2B0H7 A0A182JGI0 A0A182VN68 A0A182TXN6 A0A2R5LNQ0 A0A0K8TSF9 A0A131YKX4 T1HLF6 A0A182NRH5 R4G8X9 A0A224Z2H8 A0A336M097 L7M851 U5EXI2 A0A023FX51 A0A182TAR1 A0A131Y076 A0A026X2T0 A0A182FT11 B7Q4B3 A0A1A9X1T7 A0A1W4WS05 W5JUZ3 A0A182Y6I7 A0A195CYG6 A0A195EHU9 A0A069DU53 A0A1L8DRX1 A0A1E1XCL4 A0A0V0G9C8 A0A2R7X2A3 D3TPV7 F4W8F9 A0A023F915 A0A1A9Z6X6 A0A0L0CFA3 A0A1I8PR30 A0A1B0BXD2 E2BW48 A0A158NK65 A0A1Z5KV19 A0A1A9YBV0 A0A1A9US60 A0A151X6P4 A0A232ELV3 A0A0K8UI83 A0A1B6KQW0 A0A0K8V0J5 A0A1I8NDD8 A0A023GKF0 G3MLP9 A0A293MQG1 W8BNP5 E9H6E9 A0A0A9Y0H3 B4KCB6 A0A034W977 K7J2Z4 A0A0M3QX99 A0A0P5PRQ2 A0A0P5JIZ7 A0A0A1WVK8 A0A0P5QU46 A0A0P5CF70 A0A0P5PAC4 A0A0P5M6M5 A0A0P5RU68 A0A0P5QZG6 A0A0P5VF14 W4YX98 B3LWE9 B4LZ70 A0A1B6DJ94 B4JUH0 A0A0L7RHD8
PDB
1OU5
E-value=1.48109e-107,
Score=995
Ontologies
GO
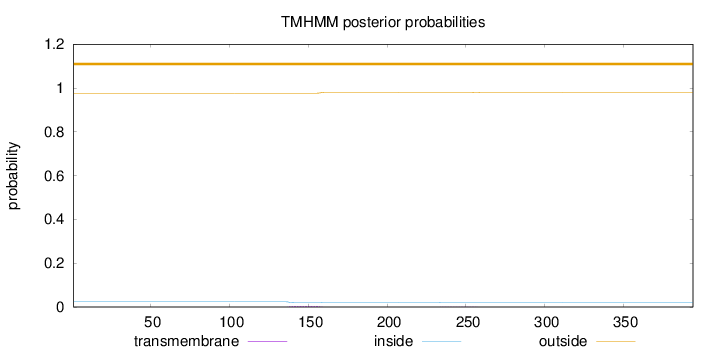
Topology
Length:
394
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0903200000000001
Exp number, first 60 AAs:
0.00037
Total prob of N-in:
0.02363
outside
1 - 394
Population Genetic Test Statistics
Pi
258.487896
Theta
213.884886
Tajima's D
0.650689
CLR
0.430906
CSRT
0.560421978901055
Interpretation
Uncertain
