Gene
KWMTBOMO15119
Annotation
PREDICTED:_uncharacterized_protein_LOC106130647_[Amyelois_transitella]
Full name
Abscission/NoCut checkpoint regulator
Alternative Name
MLL partner containing FYVE domain
Zinc finger FYVE domain-containing protein 19
Zinc finger FYVE domain-containing protein 19
Location in the cell
Nuclear Reliability : 4.194
Sequence
CDS
ATGGCGTGTAATACTTGTGCGAAACCTTTCAATTTACTGCGGAAAGAAAAGGGATGTCCAGGTTGTGGATTTAGTTATTGTTCAAAATGTCTTGACCACAAAATGTTTTTGGAGAAACTGAATGCTGAAGCAAAGGTATGTGTACAATGTAAAAAAAAATCTAATTATGATAAAAAAATAGAGCCTCCATCTGCTTACTTCAAAAGATTAGCTACTGTTCAACCACCAGATCCAAATGATAGCAATACAAACTATGCTGGAGCAAGTTCAAAAGACAATGAGATTTTAAAACGATTACAGAACCTAAAGCAAGAACAAGTTCACAAGAAAGTTTCCACAGATGACGAGATTGCTCAGAGGCTTAAAAATATAAAAGGTGACCTTCCATCAGCAAGTGTCAGTGAAATTGAAGCACGACTGGCTAATTTAAGAGGTGTACCTGTACAATCACATTGTAAGCCCATATTACCCACTCCAGATTTAAGAACGGAACAGGAGCAGGCAGATGATTTGATGAAACAGTATTTGGAGCAAGTTAATATAGACACAAAATATAAAGATGATTTTAATGATATAATAGACAGTATTGAATCTAGAGTGCAAAAATTGAAATCATCAAGTACCGAGATATCAGAAGATAAAAGTAAATTATTGTCAAATAATAATACAGAATTAGATGAAGAAGAAACAGTGAATAAAATAATTGAAAAGATAAAGGCTGATCCTTCACTTGATGAAACAGTAGATTCACCTACAAAAAATGATGAGTTGCCTTTTTGTGAGATATGTAATGAAGATGCAGTATTGCGGTGTTTGGGATGTCGATATTTGTTTTGTAAGCTTTGCTTCATGGAACACAAAGATGATGATGATGGCTGTGATAGATACGAAAATTATCAACCTCCAACAAATTAA
Protein
MACNTCAKPFNLLRKEKGCPGCGFSYCSKCLDHKMFLEKLNAEAKVCVQCKKKSNYDKKIEPPSAYFKRLATVQPPDPNDSNTNYAGASSKDNEILKRLQNLKQEQVHKKVSTDDEIAQRLKNIKGDLPSASVSEIEARLANLRGVPVQSHCKPILPTPDLRTEQEQADDLMKQYLEQVNIDTKYKDDFNDIIDSIESRVQKLKSSSTEISEDKSKLLSNNNTELDEEETVNKIIEKIKADPSLDETVDSPTKNDELPFCEICNEDAVLRCLGCRYLFCKLCFMEHKDDDDGCDRYENYQPPTN
Summary
Description
Key regulator of abscission step in cytokinesis: part of the cytokinesis checkpoint, a process required to delay abscission to prevent both premature resolution of intercellular chromosome bridges and accumulation of DNA damage. Together with CHMP4C, required to retain abscission-competent VPS4 (VPS4A and/or VPS4B) at the midbody ring until abscission checkpoint signaling is terminated at late cytokinesis. Deactivation of AURKB results in dephosphorylation of CHMP4C followed by its dissociation from ZFYVE19/ANCHR and VPS4 and subsequent abscission.
Key regulator of abscission step in cytokinesis: part of the cytokinesis checkpoint, a process required to delay abscission to prevent both premature resolution of intercellular chromosome bridges and accumulation of DNA damage. Together with CHMP4C, required to retain abscission-competent VPS4 (VPS4A and/or VPS4B) at the midbody ring until abscission checkpoint signaling is terminated at late cytokinesis. Deactivation of AURKB results in dephosphorylation of CHMP4C followed by its dissociation from ZFYVE19/ANCHR and VPS4 and subsequent abscission (By similarity).
Key regulator of abscission step in cytokinesis: part of the cytokinesis checkpoint, a process required to delay abscission to prevent both premature resolution of intercellular chromosome bridges and accumulation of DNA damage. Together with CHMP4C, required to retain abscission-competent VPS4 (VPS4A and/or VPS4B) at the midbody ring until abscission checkpoint signaling is terminated at late cytokinesis. Deactivation of AURKB results in dephosphorylation of CHMP4C followed by its dissociation from ZFYVE19/ANCHR and VPS4 and subsequent abscission (By similarity).
Subunit
Interacts (via MIM1-B) with VPS4A; interaction takes place at the midbody ring following cytokinesis checkpoint activation.
Keywords
Alternative splicing
Cell cycle
Cell division
Chromosomal rearrangement
Coiled coil
Complete proteome
Cytoplasm
Cytoskeleton
Isopeptide bond
Lipid-binding
Metal-binding
Phosphoprotein
Polymorphism
Proto-oncogene
Reference proteome
Ubl conjugation
Zinc
Zinc-finger
3D-structure
Feature
chain Abscission/NoCut checkpoint regulator
splice variant In isoform 4.
sequence variant In dbSNP:rs34819163.
splice variant In isoform 4.
sequence variant In dbSNP:rs34819163.
Uniprot
A0A2H1W1Q7
A0A212FKX2
A0A0L7KS53
A0A194PMA6
S4PHU8
A0A194RL26
+ More
A0A1I8MPZ9 A0A2I3SR96 A0A2R9BSX9 A0A2I2ZHW8 A0A0K8TS31 Q96K21-3 Q9DAZ9 Q96K21 H3BRF9 A0A2K5VEY2 F7ISY7 A0A2K6UBD3 A0A2R8P4A1 H2Q970 A0A2R9BPE8 G3QCV4 A0A2R9BQ49 A0A2I3SJ48 F7FW77 A0A2K6NQZ0 A0A2K5XF75 A0A2K5LEC3 A0A2K5XF77 A0A2K6KW02 A0A2K5H9S4 A0A1J1J0V0 A0A2K6NQY4 A0A2K6NQY7 A0A1I8Q5D5 A0A2K5LE63 A0A2K5XF74 A0A2K5H9R4 A0A2K6KVZ8 Q499R6 G3RN38 A0A2K5VER2 A0A2K5H9Q0 A0A2K5VEZ4 A0A2K6NQY2 A0A2K6B434 A0A2K5LE58 A0A2K6KVZ0 F7ISZ0 A0A2J8S5Z7 A0A1D5Q4G3 G1Q682 A0A1D5QCI0 A0A1D5QII7 B4PX72 S7P8D9 A0A2J8S600 H0VQA1 A0A2K6B424 A0A2K6B426 A0A1Q3F6V8 A0A3B0JWM5 G3TDS0 K7G4H8 A0A0D9R5F0 H2NMV4 A0A1A6H4T0 A0A1A9W8I6 G3HYR7 A0A0L0C248 G1SP94 A0A1Q3F6N2 B3NXJ7 A0A2K5S8L5 A0A2K6UBF1 A0A2K6UBD0 A0A2K5S8N7 A0A2K6GMT9 A0A2K5S8P9 A0A384D135 A0A182I006 A0A182V6Q1 A0A2K5EVE8 A0A3B3RJK4 A0A1S3AN04 A0A3Q0DIK3 A0A3B3RKC4 A0A0A1WPM9 B4MGD7 A0A2K5S8N3 Q7PUI4 A0A182X0B6 Q4QQ70 A0A1L8DWA1 T1DH90 A0A1W7RBF4 K7G4F6 F1QWA6 A8E531
A0A1I8MPZ9 A0A2I3SR96 A0A2R9BSX9 A0A2I2ZHW8 A0A0K8TS31 Q96K21-3 Q9DAZ9 Q96K21 H3BRF9 A0A2K5VEY2 F7ISY7 A0A2K6UBD3 A0A2R8P4A1 H2Q970 A0A2R9BPE8 G3QCV4 A0A2R9BQ49 A0A2I3SJ48 F7FW77 A0A2K6NQZ0 A0A2K5XF75 A0A2K5LEC3 A0A2K5XF77 A0A2K6KW02 A0A2K5H9S4 A0A1J1J0V0 A0A2K6NQY4 A0A2K6NQY7 A0A1I8Q5D5 A0A2K5LE63 A0A2K5XF74 A0A2K5H9R4 A0A2K6KVZ8 Q499R6 G3RN38 A0A2K5VER2 A0A2K5H9Q0 A0A2K5VEZ4 A0A2K6NQY2 A0A2K6B434 A0A2K5LE58 A0A2K6KVZ0 F7ISZ0 A0A2J8S5Z7 A0A1D5Q4G3 G1Q682 A0A1D5QCI0 A0A1D5QII7 B4PX72 S7P8D9 A0A2J8S600 H0VQA1 A0A2K6B424 A0A2K6B426 A0A1Q3F6V8 A0A3B0JWM5 G3TDS0 K7G4H8 A0A0D9R5F0 H2NMV4 A0A1A6H4T0 A0A1A9W8I6 G3HYR7 A0A0L0C248 G1SP94 A0A1Q3F6N2 B3NXJ7 A0A2K5S8L5 A0A2K6UBF1 A0A2K6UBD0 A0A2K5S8N7 A0A2K6GMT9 A0A2K5S8P9 A0A384D135 A0A182I006 A0A182V6Q1 A0A2K5EVE8 A0A3B3RJK4 A0A1S3AN04 A0A3Q0DIK3 A0A3B3RKC4 A0A0A1WPM9 B4MGD7 A0A2K5S8N3 Q7PUI4 A0A182X0B6 Q4QQ70 A0A1L8DWA1 T1DH90 A0A1W7RBF4 K7G4F6 F1QWA6 A8E531
Pubmed
22118469
26227816
26354079
23622113
25315136
16136131
+ More
22722832 22398555 26369729 12618766 14702039 16572171 15489334 19367720 18669648 18318008 19690332 20068231 21406692 23186163 24275569 24814515 25218447 28112733 16141072 19468303 17242355 18630941 19144319 19131326 21183079 17431167 25362486 15057822 15632090 22673903 21993624 17994087 17550304 17381049 21804562 26108605 24813606 29240929 25830018 12364791 14747013 17210077 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 23758969 26358130 23594743 12477932
22722832 22398555 26369729 12618766 14702039 16572171 15489334 19367720 18669648 18318008 19690332 20068231 21406692 23186163 24275569 24814515 25218447 28112733 16141072 19468303 17242355 18630941 19144319 19131326 21183079 17431167 25362486 15057822 15632090 22673903 21993624 17994087 17550304 17381049 21804562 26108605 24813606 29240929 25830018 12364791 14747013 17210077 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 23758969 26358130 23594743 12477932
EMBL
ODYU01005792
SOQ47025.1
AGBW02007949
OWR54393.1
JTDY01006327
KOB66103.1
+ More
KQ459604 KPI92250.1 GAIX01000354 JAA92206.1 KQ460045 KPJ18247.1 AACZ04038081 NBAG03000046 PNI94386.1 AJFE02093133 CABD030095576 GDAI01000645 JAI16958.1 AF445414 AK027746 AK122779 AC012476 CH471125 BC021092 AK005386 AL772264 AL929318 BC018420 AQIA01062298 PNI94383.1 PNI94381.1 JSUE03038066 CVRI01000066 CRL06023.1 AC111293 BC099795 CH473949 AAH99795.1 EDL79896.1 NDHI03003606 PNJ16193.1 AAPE02025705 CM000162 EDX01835.2 KE161462 EPQ03977.1 PNJ16195.1 AAKN02015886 GFDL01011744 JAV23301.1 OUUW01000011 SPP86487.1 AGCU01123730 AGCU01123731 AQIB01116308 ABGA01056823 PNJ16191.1 LZPO01047449 OBS73331.1 JH000944 EGW09307.1 JRES01000996 KNC26322.1 AAGW02024521 GFDL01011872 JAV23173.1 CH954180 EDV46956.2 APCN01002028 GBXI01013939 JAD00353.1 CH940672 EDW57460.1 AAAB01008987 EAA01452.3 AE014298 BT023531 BT023546 AAF48518.2 AAY84931.1 AAY84946.1 GFDF01003376 JAV10708.1 GAAZ01003003 JAA94940.1 GDAY02002991 JAV48476.1 CABZ01043842 CABZ01052589 CABZ01052590 BC153443 AAI53444.1
KQ459604 KPI92250.1 GAIX01000354 JAA92206.1 KQ460045 KPJ18247.1 AACZ04038081 NBAG03000046 PNI94386.1 AJFE02093133 CABD030095576 GDAI01000645 JAI16958.1 AF445414 AK027746 AK122779 AC012476 CH471125 BC021092 AK005386 AL772264 AL929318 BC018420 AQIA01062298 PNI94383.1 PNI94381.1 JSUE03038066 CVRI01000066 CRL06023.1 AC111293 BC099795 CH473949 AAH99795.1 EDL79896.1 NDHI03003606 PNJ16193.1 AAPE02025705 CM000162 EDX01835.2 KE161462 EPQ03977.1 PNJ16195.1 AAKN02015886 GFDL01011744 JAV23301.1 OUUW01000011 SPP86487.1 AGCU01123730 AGCU01123731 AQIB01116308 ABGA01056823 PNJ16191.1 LZPO01047449 OBS73331.1 JH000944 EGW09307.1 JRES01000996 KNC26322.1 AAGW02024521 GFDL01011872 JAV23173.1 CH954180 EDV46956.2 APCN01002028 GBXI01013939 JAD00353.1 CH940672 EDW57460.1 AAAB01008987 EAA01452.3 AE014298 BT023531 BT023546 AAF48518.2 AAY84931.1 AAY84946.1 GFDF01003376 JAV10708.1 GAAZ01003003 JAA94940.1 GDAY02002991 JAV48476.1 CABZ01043842 CABZ01052589 CABZ01052590 BC153443 AAI53444.1
Proteomes
UP000007151
UP000037510
UP000053268
UP000053240
UP000095301
UP000002277
+ More
UP000240080 UP000001519 UP000005640 UP000000589 UP000233100 UP000008225 UP000233220 UP000006718 UP000233200 UP000233140 UP000233060 UP000233180 UP000233080 UP000183832 UP000095300 UP000002494 UP000233120 UP000001074 UP000002282 UP000005447 UP000268350 UP000007646 UP000007267 UP000029965 UP000001595 UP000092124 UP000091820 UP000001075 UP000037069 UP000001811 UP000008711 UP000233040 UP000233160 UP000261680 UP000291021 UP000075840 UP000075903 UP000233020 UP000261540 UP000079721 UP000189704 UP000008792 UP000007062 UP000076407 UP000000803 UP000000437
UP000240080 UP000001519 UP000005640 UP000000589 UP000233100 UP000008225 UP000233220 UP000006718 UP000233200 UP000233140 UP000233060 UP000233180 UP000233080 UP000183832 UP000095300 UP000002494 UP000233120 UP000001074 UP000002282 UP000005447 UP000268350 UP000007646 UP000007267 UP000029965 UP000001595 UP000092124 UP000091820 UP000001075 UP000037069 UP000001811 UP000008711 UP000233040 UP000233160 UP000261680 UP000291021 UP000075840 UP000075903 UP000233020 UP000261540 UP000079721 UP000189704 UP000008792 UP000007062 UP000076407 UP000000803 UP000000437
Interpro
SUPFAM
SSF57903
SSF57903
Gene 3D
ProteinModelPortal
A0A2H1W1Q7
A0A212FKX2
A0A0L7KS53
A0A194PMA6
S4PHU8
A0A194RL26
+ More
A0A1I8MPZ9 A0A2I3SR96 A0A2R9BSX9 A0A2I2ZHW8 A0A0K8TS31 Q96K21-3 Q9DAZ9 Q96K21 H3BRF9 A0A2K5VEY2 F7ISY7 A0A2K6UBD3 A0A2R8P4A1 H2Q970 A0A2R9BPE8 G3QCV4 A0A2R9BQ49 A0A2I3SJ48 F7FW77 A0A2K6NQZ0 A0A2K5XF75 A0A2K5LEC3 A0A2K5XF77 A0A2K6KW02 A0A2K5H9S4 A0A1J1J0V0 A0A2K6NQY4 A0A2K6NQY7 A0A1I8Q5D5 A0A2K5LE63 A0A2K5XF74 A0A2K5H9R4 A0A2K6KVZ8 Q499R6 G3RN38 A0A2K5VER2 A0A2K5H9Q0 A0A2K5VEZ4 A0A2K6NQY2 A0A2K6B434 A0A2K5LE58 A0A2K6KVZ0 F7ISZ0 A0A2J8S5Z7 A0A1D5Q4G3 G1Q682 A0A1D5QCI0 A0A1D5QII7 B4PX72 S7P8D9 A0A2J8S600 H0VQA1 A0A2K6B424 A0A2K6B426 A0A1Q3F6V8 A0A3B0JWM5 G3TDS0 K7G4H8 A0A0D9R5F0 H2NMV4 A0A1A6H4T0 A0A1A9W8I6 G3HYR7 A0A0L0C248 G1SP94 A0A1Q3F6N2 B3NXJ7 A0A2K5S8L5 A0A2K6UBF1 A0A2K6UBD0 A0A2K5S8N7 A0A2K6GMT9 A0A2K5S8P9 A0A384D135 A0A182I006 A0A182V6Q1 A0A2K5EVE8 A0A3B3RJK4 A0A1S3AN04 A0A3Q0DIK3 A0A3B3RKC4 A0A0A1WPM9 B4MGD7 A0A2K5S8N3 Q7PUI4 A0A182X0B6 Q4QQ70 A0A1L8DWA1 T1DH90 A0A1W7RBF4 K7G4F6 F1QWA6 A8E531
A0A1I8MPZ9 A0A2I3SR96 A0A2R9BSX9 A0A2I2ZHW8 A0A0K8TS31 Q96K21-3 Q9DAZ9 Q96K21 H3BRF9 A0A2K5VEY2 F7ISY7 A0A2K6UBD3 A0A2R8P4A1 H2Q970 A0A2R9BPE8 G3QCV4 A0A2R9BQ49 A0A2I3SJ48 F7FW77 A0A2K6NQZ0 A0A2K5XF75 A0A2K5LEC3 A0A2K5XF77 A0A2K6KW02 A0A2K5H9S4 A0A1J1J0V0 A0A2K6NQY4 A0A2K6NQY7 A0A1I8Q5D5 A0A2K5LE63 A0A2K5XF74 A0A2K5H9R4 A0A2K6KVZ8 Q499R6 G3RN38 A0A2K5VER2 A0A2K5H9Q0 A0A2K5VEZ4 A0A2K6NQY2 A0A2K6B434 A0A2K5LE58 A0A2K6KVZ0 F7ISZ0 A0A2J8S5Z7 A0A1D5Q4G3 G1Q682 A0A1D5QCI0 A0A1D5QII7 B4PX72 S7P8D9 A0A2J8S600 H0VQA1 A0A2K6B424 A0A2K6B426 A0A1Q3F6V8 A0A3B0JWM5 G3TDS0 K7G4H8 A0A0D9R5F0 H2NMV4 A0A1A6H4T0 A0A1A9W8I6 G3HYR7 A0A0L0C248 G1SP94 A0A1Q3F6N2 B3NXJ7 A0A2K5S8L5 A0A2K6UBF1 A0A2K6UBD0 A0A2K5S8N7 A0A2K6GMT9 A0A2K5S8P9 A0A384D135 A0A182I006 A0A182V6Q1 A0A2K5EVE8 A0A3B3RJK4 A0A1S3AN04 A0A3Q0DIK3 A0A3B3RKC4 A0A0A1WPM9 B4MGD7 A0A2K5S8N3 Q7PUI4 A0A182X0B6 Q4QQ70 A0A1L8DWA1 T1DH90 A0A1W7RBF4 K7G4F6 F1QWA6 A8E531
PDB
1WFK
E-value=7.71389e-05,
Score=108
Ontologies
GO
Topology
Subcellular location
Cytoplasm
Localizes mainly on centrosomes in interphase and early mitosis. Localizes at the cleavage furrow and midbody ring in late mitosis and cytokinesis. With evidence from 16 publications.
Cytoskeleton Localizes mainly on centrosomes in interphase and early mitosis. Localizes at the cleavage furrow and midbody ring in late mitosis and cytokinesis. With evidence from 16 publications.
Microtubule organizing center Localizes mainly on centrosomes in interphase and early mitosis. Localizes at the cleavage furrow and midbody ring in late mitosis and cytokinesis. With evidence from 16 publications.
Centrosome Localizes mainly on centrosomes in interphase and early mitosis. Localizes at the cleavage furrow and midbody ring in late mitosis and cytokinesis. With evidence from 16 publications.
Cleavage furrow Localizes mainly on centrosomes in interphase and early mitosis. Localizes at the cleavage furrow and midbody ring in late mitosis and cytokinesis. With evidence from 16 publications.
Midbody Localizes mainly on centrosomes in interphase and early mitosis. Localizes at the cleavage furrow and midbody ring in late mitosis and cytokinesis. With evidence from 16 publications.
Midbody ring Localizes mainly on centrosomes in interphase and early mitosis. Localizes at the cleavage furrow and midbody ring in late mitosis and cytokinesis. With evidence from 16 publications.
Cytoskeleton Localizes mainly on centrosomes in interphase and early mitosis. Localizes at the cleavage furrow and midbody ring in late mitosis and cytokinesis. With evidence from 16 publications.
Microtubule organizing center Localizes mainly on centrosomes in interphase and early mitosis. Localizes at the cleavage furrow and midbody ring in late mitosis and cytokinesis. With evidence from 16 publications.
Centrosome Localizes mainly on centrosomes in interphase and early mitosis. Localizes at the cleavage furrow and midbody ring in late mitosis and cytokinesis. With evidence from 16 publications.
Cleavage furrow Localizes mainly on centrosomes in interphase and early mitosis. Localizes at the cleavage furrow and midbody ring in late mitosis and cytokinesis. With evidence from 16 publications.
Midbody Localizes mainly on centrosomes in interphase and early mitosis. Localizes at the cleavage furrow and midbody ring in late mitosis and cytokinesis. With evidence from 16 publications.
Midbody ring Localizes mainly on centrosomes in interphase and early mitosis. Localizes at the cleavage furrow and midbody ring in late mitosis and cytokinesis. With evidence from 16 publications.
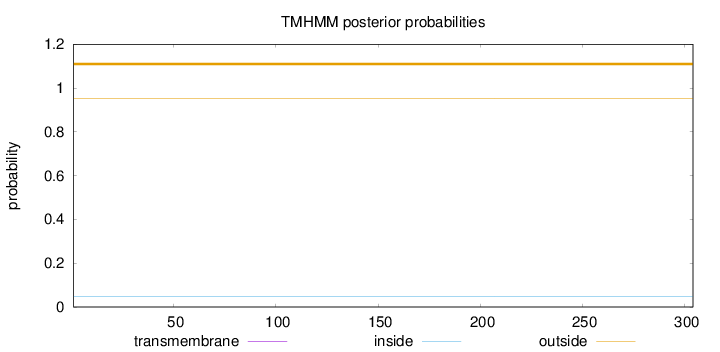
Length:
304
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.04787
outside
1 - 304
Population Genetic Test Statistics
Pi
303.454399
Theta
232.226877
Tajima's D
1.116643
CLR
0.116201
CSRT
0.690615469226539
Interpretation
Uncertain
