Gene
KWMTBOMO15115
Pre Gene Modal
BGIBMGA004995
Annotation
PREDICTED:_serine_racemase-like_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.558
Sequence
CDS
ATGATGAAGGCACTTCAGGCTGGTAAACCAGTCTCCGTGCCGATCGTCTCGAATCTTGCAGAGGGACTCAACGCTTCTAGAGTTGGAGACAATACTTTTGCTACTATTAAAGGACGACTTGATAGAATGTTGGTAGTTGAAGAGTCATATATCGCCCGTGCTATGATATCAGTATTAGAACGGGAACGGCTTGTAACGGACTGCGCTGGAGTCTGTGCTGTAGCAGCTGTCATGCAAGGTTTAGTACCAGAGCTCCGTGGAAAACGGGCGGTATGTGTGCTGAGCAGTGGAAACGTAGACTCGGGTCGCCTGTCTCGTTCTATACAACGTGGGCTCGGTGCTAGCGGTAGACTCCTTCGGTTTGCGGTTGCCGTGCCCGACCATCGATCGGGACTGGAACACCTGGCTAAAACGATCGCAAATGAGAATGCCGTTCTGAAGAGTCTTGTTACTGAGCAAATGTGGGTGCACAGCGATGTTGGCTCTACTTGGGTGAATGTTGTGGTAGAGACTGCCAACGAGGAACATGCCAATAAATTAAAGGATCGTTTGCGTGATCTTTATCCGTCTGCTAGATTTGCTGTCATTGATCTCGATGACAAATACAAAATTGCTGTGCGATAA
Protein
MMKALQAGKPVSVPIVSNLAEGLNASRVGDNTFATIKGRLDRMLVVEESYIARAMISVLERERLVTDCAGVCAVAAVMQGLVPELRGKRAVCVLSSGNVDSGRLSRSIQRGLGASGRLLRFAVAVPDHRSGLEHLAKTIANENAVLKSLVTEQMWVHSDVGSTWVNVVVETANEEHANKLKDRLRDLYPSARFAVIDLDDKYKIAVR
Summary
Uniprot
A0A0L7L2P7
A0A2H1WD40
A0A3S2L227
A0A212FKY5
A0A194RLF8
A0A194PFT2
+ More
T1I4J3 A0A224XGR1 A0A146L135 A0A0A9XZJ6 A0A0A9XU89 R7UXC2 A0A210PEA5 K1PNT5 A0A1B6I1Q5 T1FMN2 A0A0D2WRW7 A0A2A4J248 A0A3S2M3E5 E9GBI8 V3ZXV2 C3Y9T1 A0A0P5GZH8 A0A0P4WHL5 A0A2H1W1X4 A0A0P4ZTQ6 A0A0P6F9A1 A0A0P5ES17 A0A0P5BXF9 A0A0P5J6M1 A0A0P5AA51 A0A154P355 A0A162DCS6 A0A0P6ABD4 E2C2A7 A0A0L7R1C7 A0A0P6JSY7 A0A1B6FMG6 A0A016W1Y9 A0A088A0I6 H9J5W7 A0A016VZJ7 A0A0M8ZSV0 A0A212FKW8 A0A016W034 A0A3S1BB83 A0A1Y1L4P9 A0A1Y1L250 A0A0K2TDQ9 U4UCZ3 A0A2A3EQU8 T1G187 A0A1I7UDI0 A0A1D1VA39 A0A368H631 N6T538 A0A016U5Z4 A0A016U500 A0A016U3Z4 A7RKN0 A0A0V0XQK4 A0A2A2J672 A0A0V1ESN8 F0WTS3 A0A0V1HP28 A0A1I8A2A3 A0A0V1M2V2 A0A2A6C7P2 H3FW53 A0A2B4STK8 A0A0V0RIN1 K3X242 A0A024FT24 G0NFJ0 A0A0V0W658 A0A2A2LPE4 A0A1Y3ECS9 A0A2H2IKI5 D6WSN2 A0A0C2GNE9 A0A088A0I7 A0A1S3JU28 A0A2G5UXY2 A8WNC1 A0A0V1LIR7 Q21080 W2T5H7 A0A1V9YG80 A0A3P6HGR8 A0A0R3U9S2 A0A2C9JEY1 A0A194RQE8 A0A261C149 E3LGP2 A0A194PFT7 A0A261B0I5 H3G667 F2UJF1 A0A0L0CQY9
T1I4J3 A0A224XGR1 A0A146L135 A0A0A9XZJ6 A0A0A9XU89 R7UXC2 A0A210PEA5 K1PNT5 A0A1B6I1Q5 T1FMN2 A0A0D2WRW7 A0A2A4J248 A0A3S2M3E5 E9GBI8 V3ZXV2 C3Y9T1 A0A0P5GZH8 A0A0P4WHL5 A0A2H1W1X4 A0A0P4ZTQ6 A0A0P6F9A1 A0A0P5ES17 A0A0P5BXF9 A0A0P5J6M1 A0A0P5AA51 A0A154P355 A0A162DCS6 A0A0P6ABD4 E2C2A7 A0A0L7R1C7 A0A0P6JSY7 A0A1B6FMG6 A0A016W1Y9 A0A088A0I6 H9J5W7 A0A016VZJ7 A0A0M8ZSV0 A0A212FKW8 A0A016W034 A0A3S1BB83 A0A1Y1L4P9 A0A1Y1L250 A0A0K2TDQ9 U4UCZ3 A0A2A3EQU8 T1G187 A0A1I7UDI0 A0A1D1VA39 A0A368H631 N6T538 A0A016U5Z4 A0A016U500 A0A016U3Z4 A7RKN0 A0A0V0XQK4 A0A2A2J672 A0A0V1ESN8 F0WTS3 A0A0V1HP28 A0A1I8A2A3 A0A0V1M2V2 A0A2A6C7P2 H3FW53 A0A2B4STK8 A0A0V0RIN1 K3X242 A0A024FT24 G0NFJ0 A0A0V0W658 A0A2A2LPE4 A0A1Y3ECS9 A0A2H2IKI5 D6WSN2 A0A0C2GNE9 A0A088A0I7 A0A1S3JU28 A0A2G5UXY2 A8WNC1 A0A0V1LIR7 Q21080 W2T5H7 A0A1V9YG80 A0A3P6HGR8 A0A0R3U9S2 A0A2C9JEY1 A0A194RQE8 A0A261C149 E3LGP2 A0A194PFT7 A0A261B0I5 H3G667 F2UJF1 A0A0L0CQY9
Pubmed
EMBL
JTDY01003307
KOB69768.1
ODYU01007835
SOQ50947.1
RSAL01000228
RVE43913.1
+ More
AGBW02007949 OWR54397.1 KQ460045 KPJ18254.1 KQ459604 KPI92241.1 ACPB03007717 ACPB03007718 GFTR01006212 JAW10214.1 GDHC01017672 JAQ00957.1 GBHO01019326 GBRD01016356 GDHC01009780 JAG24278.1 JAG49470.1 JAQ08849.1 GBHO01019327 GBRD01016355 JAG24277.1 JAG49471.1 AMQN01005886 KB297068 ELU10999.1 NEDP02076749 OWF34823.1 JH816681 EKC18150.1 GECU01026854 GECU01001825 JAS80852.1 JAT05882.1 AMQM01002844 KB095905 ESO09945.1 KE346367 KJE94790.1 NWSH01004124 PCG65472.1 RSAL01000053 RVE50057.1 GL732538 EFX82963.1 KB202619 ESO89242.1 GG666493 EEN62840.1 GDIQ01240834 JAK10891.1 GDRN01060339 GDRN01060336 GDRN01060335 JAI65380.1 ODYU01005792 SOQ47023.1 GDIP01208678 JAJ14724.1 GDIQ01052810 JAN41927.1 GDIP01144174 JAJ79228.1 GDIP01179084 JAJ44318.1 GDIQ01229145 JAK22580.1 GDIP01214368 JAJ09034.1 KQ434809 KZC06262.1 LRGB01002860 KZS05865.1 GDIP01031947 JAM71768.1 GL452104 EFN77927.1 KQ414667 KOC64675.1 GDIQ01009675 JAN85062.1 GECZ01018364 JAS51405.1 JARK01001338 EYC32998.1 BABH01030008 BABH01030009 BABH01030010 BABH01030011 BABH01030012 EYC32999.1 KQ435863 KOX70430.1 OWR54395.1 EYC33000.1 RQTK01000454 RUS79394.1 GEZM01066589 JAV67778.1 GEZM01066588 JAV67779.1 HACA01006230 CDW23591.1 KB632263 ERL90907.1 KZ288194 PBC33874.1 ESO09946.1 BDGG01000004 GAU98519.1 JOJR01000009 RCN52061.1 APGK01052218 APGK01052219 KB741211 ENN72773.1 JARK01001394 EYC10003.1 EYC10006.1 EYC10004.1 DS469516 EDO47936.1 JYDU01000172 KRX90231.1 LIAE01010657 PAV57125.1 JYDR01000010 JYDR01000009 JYDT01000003 JYDS01000003 JYDV01000003 KRY76763.1 KRY77061.1 KRY93260.1 KRZ34484.1 KRZ45454.1 FR824302 CCA24766.1 JYDP01000050 KRZ11381.1 JYDO01000269 KRZ66067.1 ABKE03000060 PDM74179.1 LSMT01000020 PFX32676.1 JYDL01000163 KRX14345.1 GL376562 CAIX01000096 CCI10131.1 GL379876 EGT59622.1 JYDK01000245 KRX71350.1 LIAE01006533 PAV88111.1 LVZM01022133 OUC40928.1 KQ971354 EFA05892.1 KN730814 KIH60604.1 PDUG01000002 PIC44358.1 HE600999 CAP21975.1 JYDW01000047 KRZ59106.1 BX284602 CAA88854.2 KI660225 ETN76446.1 JNBS01003942 OQR84708.1 UXSR01000919 VDD77668.1 KPJ18251.1 NIPN01000047 OZG16277.1 DS268408 LFJK02001013 EFO86191.1 POM34577.1 KPI92246.1 NMWX01000004 OZG03205.1 DS566019 GL832977 EGD77250.1 JRES01000145 KNC33844.1
AGBW02007949 OWR54397.1 KQ460045 KPJ18254.1 KQ459604 KPI92241.1 ACPB03007717 ACPB03007718 GFTR01006212 JAW10214.1 GDHC01017672 JAQ00957.1 GBHO01019326 GBRD01016356 GDHC01009780 JAG24278.1 JAG49470.1 JAQ08849.1 GBHO01019327 GBRD01016355 JAG24277.1 JAG49471.1 AMQN01005886 KB297068 ELU10999.1 NEDP02076749 OWF34823.1 JH816681 EKC18150.1 GECU01026854 GECU01001825 JAS80852.1 JAT05882.1 AMQM01002844 KB095905 ESO09945.1 KE346367 KJE94790.1 NWSH01004124 PCG65472.1 RSAL01000053 RVE50057.1 GL732538 EFX82963.1 KB202619 ESO89242.1 GG666493 EEN62840.1 GDIQ01240834 JAK10891.1 GDRN01060339 GDRN01060336 GDRN01060335 JAI65380.1 ODYU01005792 SOQ47023.1 GDIP01208678 JAJ14724.1 GDIQ01052810 JAN41927.1 GDIP01144174 JAJ79228.1 GDIP01179084 JAJ44318.1 GDIQ01229145 JAK22580.1 GDIP01214368 JAJ09034.1 KQ434809 KZC06262.1 LRGB01002860 KZS05865.1 GDIP01031947 JAM71768.1 GL452104 EFN77927.1 KQ414667 KOC64675.1 GDIQ01009675 JAN85062.1 GECZ01018364 JAS51405.1 JARK01001338 EYC32998.1 BABH01030008 BABH01030009 BABH01030010 BABH01030011 BABH01030012 EYC32999.1 KQ435863 KOX70430.1 OWR54395.1 EYC33000.1 RQTK01000454 RUS79394.1 GEZM01066589 JAV67778.1 GEZM01066588 JAV67779.1 HACA01006230 CDW23591.1 KB632263 ERL90907.1 KZ288194 PBC33874.1 ESO09946.1 BDGG01000004 GAU98519.1 JOJR01000009 RCN52061.1 APGK01052218 APGK01052219 KB741211 ENN72773.1 JARK01001394 EYC10003.1 EYC10006.1 EYC10004.1 DS469516 EDO47936.1 JYDU01000172 KRX90231.1 LIAE01010657 PAV57125.1 JYDR01000010 JYDR01000009 JYDT01000003 JYDS01000003 JYDV01000003 KRY76763.1 KRY77061.1 KRY93260.1 KRZ34484.1 KRZ45454.1 FR824302 CCA24766.1 JYDP01000050 KRZ11381.1 JYDO01000269 KRZ66067.1 ABKE03000060 PDM74179.1 LSMT01000020 PFX32676.1 JYDL01000163 KRX14345.1 GL376562 CAIX01000096 CCI10131.1 GL379876 EGT59622.1 JYDK01000245 KRX71350.1 LIAE01006533 PAV88111.1 LVZM01022133 OUC40928.1 KQ971354 EFA05892.1 KN730814 KIH60604.1 PDUG01000002 PIC44358.1 HE600999 CAP21975.1 JYDW01000047 KRZ59106.1 BX284602 CAA88854.2 KI660225 ETN76446.1 JNBS01003942 OQR84708.1 UXSR01000919 VDD77668.1 KPJ18251.1 NIPN01000047 OZG16277.1 DS268408 LFJK02001013 EFO86191.1 POM34577.1 KPI92246.1 NMWX01000004 OZG03205.1 DS566019 GL832977 EGD77250.1 JRES01000145 KNC33844.1
Proteomes
UP000037510
UP000283053
UP000007151
UP000053240
UP000053268
UP000015103
+ More
UP000014760 UP000242188 UP000005408 UP000015101 UP000008743 UP000218220 UP000000305 UP000030746 UP000001554 UP000076502 UP000076858 UP000008237 UP000053825 UP000024635 UP000005203 UP000005204 UP000053105 UP000271974 UP000030742 UP000242457 UP000095282 UP000186922 UP000252519 UP000019118 UP000001593 UP000054815 UP000218231 UP000054632 UP000054805 UP000054826 UP000054995 UP000055024 UP000095287 UP000054843 UP000232936 UP000005239 UP000225706 UP000054630 UP000053237 UP000008068 UP000054673 UP000243006 UP000005237 UP000007266 UP000085678 UP000230233 UP000008549 UP000054721 UP000001940 UP000243217 UP000267029 UP000046399 UP000076420 UP000216463 UP000008281 UP000237256 UP000216624 UP000005238 UP000007799 UP000037069
UP000014760 UP000242188 UP000005408 UP000015101 UP000008743 UP000218220 UP000000305 UP000030746 UP000001554 UP000076502 UP000076858 UP000008237 UP000053825 UP000024635 UP000005203 UP000005204 UP000053105 UP000271974 UP000030742 UP000242457 UP000095282 UP000186922 UP000252519 UP000019118 UP000001593 UP000054815 UP000218231 UP000054632 UP000054805 UP000054826 UP000054995 UP000055024 UP000095287 UP000054843 UP000232936 UP000005239 UP000225706 UP000054630 UP000053237 UP000008068 UP000054673 UP000243006 UP000005237 UP000007266 UP000085678 UP000230233 UP000008549 UP000054721 UP000001940 UP000243217 UP000267029 UP000046399 UP000076420 UP000216463 UP000008281 UP000237256 UP000216624 UP000005238 UP000007799 UP000037069
Interpro
Gene 3D
CDD
ProteinModelPortal
A0A0L7L2P7
A0A2H1WD40
A0A3S2L227
A0A212FKY5
A0A194RLF8
A0A194PFT2
+ More
T1I4J3 A0A224XGR1 A0A146L135 A0A0A9XZJ6 A0A0A9XU89 R7UXC2 A0A210PEA5 K1PNT5 A0A1B6I1Q5 T1FMN2 A0A0D2WRW7 A0A2A4J248 A0A3S2M3E5 E9GBI8 V3ZXV2 C3Y9T1 A0A0P5GZH8 A0A0P4WHL5 A0A2H1W1X4 A0A0P4ZTQ6 A0A0P6F9A1 A0A0P5ES17 A0A0P5BXF9 A0A0P5J6M1 A0A0P5AA51 A0A154P355 A0A162DCS6 A0A0P6ABD4 E2C2A7 A0A0L7R1C7 A0A0P6JSY7 A0A1B6FMG6 A0A016W1Y9 A0A088A0I6 H9J5W7 A0A016VZJ7 A0A0M8ZSV0 A0A212FKW8 A0A016W034 A0A3S1BB83 A0A1Y1L4P9 A0A1Y1L250 A0A0K2TDQ9 U4UCZ3 A0A2A3EQU8 T1G187 A0A1I7UDI0 A0A1D1VA39 A0A368H631 N6T538 A0A016U5Z4 A0A016U500 A0A016U3Z4 A7RKN0 A0A0V0XQK4 A0A2A2J672 A0A0V1ESN8 F0WTS3 A0A0V1HP28 A0A1I8A2A3 A0A0V1M2V2 A0A2A6C7P2 H3FW53 A0A2B4STK8 A0A0V0RIN1 K3X242 A0A024FT24 G0NFJ0 A0A0V0W658 A0A2A2LPE4 A0A1Y3ECS9 A0A2H2IKI5 D6WSN2 A0A0C2GNE9 A0A088A0I7 A0A1S3JU28 A0A2G5UXY2 A8WNC1 A0A0V1LIR7 Q21080 W2T5H7 A0A1V9YG80 A0A3P6HGR8 A0A0R3U9S2 A0A2C9JEY1 A0A194RQE8 A0A261C149 E3LGP2 A0A194PFT7 A0A261B0I5 H3G667 F2UJF1 A0A0L0CQY9
T1I4J3 A0A224XGR1 A0A146L135 A0A0A9XZJ6 A0A0A9XU89 R7UXC2 A0A210PEA5 K1PNT5 A0A1B6I1Q5 T1FMN2 A0A0D2WRW7 A0A2A4J248 A0A3S2M3E5 E9GBI8 V3ZXV2 C3Y9T1 A0A0P5GZH8 A0A0P4WHL5 A0A2H1W1X4 A0A0P4ZTQ6 A0A0P6F9A1 A0A0P5ES17 A0A0P5BXF9 A0A0P5J6M1 A0A0P5AA51 A0A154P355 A0A162DCS6 A0A0P6ABD4 E2C2A7 A0A0L7R1C7 A0A0P6JSY7 A0A1B6FMG6 A0A016W1Y9 A0A088A0I6 H9J5W7 A0A016VZJ7 A0A0M8ZSV0 A0A212FKW8 A0A016W034 A0A3S1BB83 A0A1Y1L4P9 A0A1Y1L250 A0A0K2TDQ9 U4UCZ3 A0A2A3EQU8 T1G187 A0A1I7UDI0 A0A1D1VA39 A0A368H631 N6T538 A0A016U5Z4 A0A016U500 A0A016U3Z4 A7RKN0 A0A0V0XQK4 A0A2A2J672 A0A0V1ESN8 F0WTS3 A0A0V1HP28 A0A1I8A2A3 A0A0V1M2V2 A0A2A6C7P2 H3FW53 A0A2B4STK8 A0A0V0RIN1 K3X242 A0A024FT24 G0NFJ0 A0A0V0W658 A0A2A2LPE4 A0A1Y3ECS9 A0A2H2IKI5 D6WSN2 A0A0C2GNE9 A0A088A0I7 A0A1S3JU28 A0A2G5UXY2 A8WNC1 A0A0V1LIR7 Q21080 W2T5H7 A0A1V9YG80 A0A3P6HGR8 A0A0R3U9S2 A0A2C9JEY1 A0A194RQE8 A0A261C149 E3LGP2 A0A194PFT7 A0A261B0I5 H3G667 F2UJF1 A0A0L0CQY9
PDB
1TDJ
E-value=2.02976e-10,
Score=154
Ontologies
PATHWAY
00260
Glycine, serine and threonine metabolism - Bombyx mori (domestic silkworm)
00290 Valine, leucine and isoleucine biosynthesis - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
01230 Biosynthesis of amino acids - Bombyx mori (domestic silkworm)
00290 Valine, leucine and isoleucine biosynthesis - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
01230 Biosynthesis of amino acids - Bombyx mori (domestic silkworm)
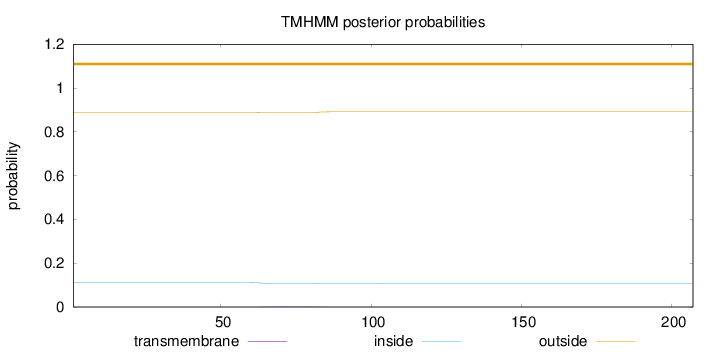
Topology
Length:
207
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0781699999999999
Exp number, first 60 AAs:
0.0017
Total prob of N-in:
0.11117
outside
1 - 207
Population Genetic Test Statistics
Pi
205.661975
Theta
194.582071
Tajima's D
0.343069
CLR
49.04067
CSRT
0.467976601169941
Interpretation
Uncertain
