Pre Gene Modal
BGIBMGA004905
Annotation
hypothetical_protein_KGM_16288_[Danaus_plexippus]
Location in the cell
Cytoplasmic Reliability : 1.146 Mitochondrial Reliability : 1.425
Sequence
CDS
ATGACTTATGCACATACGTACAAGGCTCACTCTACTTTCAGGTACAAACTAAGACCTGAAGTAAATTTGGAAGCCGTCGCAGAGGCACTACCCGACAAGTGCACCGGCGCTGACTTACTGCAGGTTGTCTCTACTGCACGGTCAGCGGCCGTGAGAGCACTCGTTGCTAAGCTACACAATGGAACGTTAAAGGAAAGCGAACTGAGTCCCGACTCCATCGTTATAGGGGTAAGTGAACTATGGGAGGGCGTTGTATCATTCCGGCCCTCGGTCACAGACGATGAACTGTCGCATTACGAAGCGATGGAGTCACAAATGTAA
Protein
MTYAHTYKAHSTFRYKLRPEVNLEAVAEALPDKCTGADLLQVVSTARSAAVRALVAKLHNGTLKESELSPDSIVIGVSELWEGVVSFRPSVTDDELSHYEAMESQM
Summary
Similarity
Belongs to the AAA ATPase family.
Uniprot
A0A212FL30
A0A194PHP2
A0A2H1WD00
A0A2J7PH22
A0A2J7PH28
A0A1B0G0P0
+ More
A0A1A9USQ0 A0A1B0BE70 A0A3B4Z067 A0A3B4Z062 A0A3Q3K0K2 A0A0L0BNF3 A0A3Q1F3X5 A0A1I8Q7G9 A0A1I8Q7F9 A0A1A8G552 A0A3Q1CWW4 A0A1A8EDS6 B4QAP6 A0A3B5LYC1 B4HN18 A0A0J9TYL5 A0A0J9RA23 A0A3B5LYB7 M3ZUW5 A0A3B5LYY1 A0A3P8TL71 A0A1A7Z2R1 A0A1A7ZJ39 B4MKB9 A0A1A8ER57 A0A3P9Q7P6 B3NN35 A0A0Q5VPL1 A0A0R1DNM7 B4P7N0
A0A1A9USQ0 A0A1B0BE70 A0A3B4Z067 A0A3B4Z062 A0A3Q3K0K2 A0A0L0BNF3 A0A3Q1F3X5 A0A1I8Q7G9 A0A1I8Q7F9 A0A1A8G552 A0A3Q1CWW4 A0A1A8EDS6 B4QAP6 A0A3B5LYC1 B4HN18 A0A0J9TYL5 A0A0J9RA23 A0A3B5LYB7 M3ZUW5 A0A3B5LYY1 A0A3P8TL71 A0A1A7Z2R1 A0A1A7ZJ39 B4MKB9 A0A1A8ER57 A0A3P9Q7P6 B3NN35 A0A0Q5VPL1 A0A0R1DNM7 B4P7N0
EMBL
AGBW02007949
OWR54399.1
KQ459604
KPI92239.1
ODYU01007835
SOQ50948.1
+ More
NEVH01025150 PNF15633.1 PNF15634.1 CCAG010023102 JXJN01012803 JRES01001601 KNC21562.1 HAEB01019706 SBQ66233.1 HAEA01015847 SBQ44328.1 CM000362 EDX06551.1 CH480816 EDW47322.1 CM002911 KMY92880.1 KMY92881.1 KMY92882.1 HADW01010125 HADX01014511 SBP36743.1 HADY01003831 HAEJ01005467 SBP42316.1 CH963846 EDW72558.1 HAEB01002825 SBQ49352.1 CH954179 EDV56555.1 KQS62900.1 CM000158 KRJ98885.1 EDW90065.1
NEVH01025150 PNF15633.1 PNF15634.1 CCAG010023102 JXJN01012803 JRES01001601 KNC21562.1 HAEB01019706 SBQ66233.1 HAEA01015847 SBQ44328.1 CM000362 EDX06551.1 CH480816 EDW47322.1 CM002911 KMY92880.1 KMY92881.1 KMY92882.1 HADW01010125 HADX01014511 SBP36743.1 HADY01003831 HAEJ01005467 SBP42316.1 CH963846 EDW72558.1 HAEB01002825 SBQ49352.1 CH954179 EDV56555.1 KQS62900.1 CM000158 KRJ98885.1 EDW90065.1
Proteomes
Interpro
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
A0A212FL30
A0A194PHP2
A0A2H1WD00
A0A2J7PH22
A0A2J7PH28
A0A1B0G0P0
+ More
A0A1A9USQ0 A0A1B0BE70 A0A3B4Z067 A0A3B4Z062 A0A3Q3K0K2 A0A0L0BNF3 A0A3Q1F3X5 A0A1I8Q7G9 A0A1I8Q7F9 A0A1A8G552 A0A3Q1CWW4 A0A1A8EDS6 B4QAP6 A0A3B5LYC1 B4HN18 A0A0J9TYL5 A0A0J9RA23 A0A3B5LYB7 M3ZUW5 A0A3B5LYY1 A0A3P8TL71 A0A1A7Z2R1 A0A1A7ZJ39 B4MKB9 A0A1A8ER57 A0A3P9Q7P6 B3NN35 A0A0Q5VPL1 A0A0R1DNM7 B4P7N0
A0A1A9USQ0 A0A1B0BE70 A0A3B4Z067 A0A3B4Z062 A0A3Q3K0K2 A0A0L0BNF3 A0A3Q1F3X5 A0A1I8Q7G9 A0A1I8Q7F9 A0A1A8G552 A0A3Q1CWW4 A0A1A8EDS6 B4QAP6 A0A3B5LYC1 B4HN18 A0A0J9TYL5 A0A0J9RA23 A0A3B5LYB7 M3ZUW5 A0A3B5LYY1 A0A3P8TL71 A0A1A7Z2R1 A0A1A7ZJ39 B4MKB9 A0A1A8ER57 A0A3P9Q7P6 B3NN35 A0A0Q5VPL1 A0A0R1DNM7 B4P7N0
Ontologies
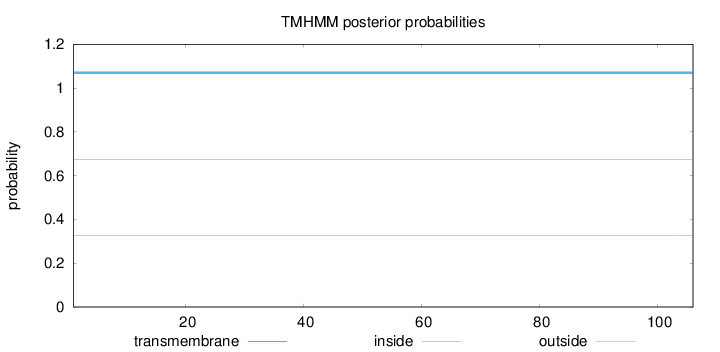
Topology
Length:
106
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0023
Exp number, first 60 AAs:
0.00154
Total prob of N-in:
0.67222
inside
1 - 106
Population Genetic Test Statistics
Pi
371.042519
Theta
210.184601
Tajima's D
2.337345
CLR
0
CSRT
0.929303534823259
Interpretation
Uncertain
