Pre Gene Modal
BGIBMGA004905
Annotation
PREDICTED:_peroxisome_biogenesis_protein_6_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.78
Sequence
CDS
ATGTGTATCACAACATTACGTGTGACCCAACTAACTTTAGGAGAGAATATTCATGCAACAAAGCCACAAGATAAATATTTCAAGCCACAAGGATTTGATGATAATTTACACATGCTAGAACTCTGCCCAACTGGCTTAAAGCATATTTTTAATCAAATTCAAGAAACCATTTCACCATTTTTAAATGAAGATTTAAGTGACCTCTCATTGTTTTTATCGAGTCCGATTATACCAATGTTAGCCTTAACCGGCTGCTTGGGGTCTGGAAGACACTTAGCTGTAAAAACATTAGCAAAATTTAATGGAATTAATTGCCTCTCTATTGATTGCAATGCATTACAAAGCTCAACAGCTAAGCAGACTGAAAGTAAAATTAATGCAACCATACAAAAGGCAAAGGCTGCGGCACCTGTCATTGTGTTACTAGATAATTTTGAGGTATTTGCAATAGATGCAGAAAACAATGAGGACTGTAGAGTGATTGAATACTTTATGAACACAATCACTGATTTATATCAGAACTACACAAAGCACCCTATCATATTCATTGCTGTGACGAAAGATATAGAATTTAAACCGAACATAAGTAGGCTGTTTCTAGAAAAGTTTCACATACCGAAATTGAATCTTAGACAGAGATATGAGATGCTAGACTGGTTTTCCAACGTAATGAGACTAAATATAGATGGTGCAGAGCATGTGAAAGCAAATGAGTTATTAGTAAAACAAACACCAGTGAATGTAAATAGTCAGAATGTTTTACAAAGGGTGGCAGCGAAGACTGAAACGTTTTTGTATGGCGATTTGGACACATTAGTGCACTTCGCGACTAGAGAGTCGTATTTGAGGCAGCACAATAATTATAATCAACTACCGCCCGATCCTGACTTGCGGTTAATAAAGGAAGAAGATTTTAACTCGGCTTTAGAATCAATAAGAAAGCTACAAAGTCAACATTTGGACGCGCCGAAAATACCCAAAGTGTACTGGGATGATATCGGAGGGTTGGAGAAACTAAAAAAGGAATTACTTAAGACAATACAGCTTCCTATTAAATACCCACACTTGTTCAAAAACGCGAGCCTCAAAAGATCAGGCATACTGTTGTACGGGCCTCCAGGTTGTGGGAAAACGTTAGTAGCTAAAGCAGTAAGCACGGAGTTGAATGTGAGCTTTCTCAGTGTCAAAGGTCCCGAGCTACTAAATATGTACATCGGTCAGTCCGAAGAGAATGTTAGAAAAGTCTTCGAGTCTGCACGCGCATCGTCTCCGTGCATAATATTCGTGGACGAGCTGGACGCGCTGGCGCCGCGCCGCGGGGCCGCCGGGGACTCCGCCGGCGCCAGCGACCGCGTCGTCAGCCAACTGCTCGCTGAACTCGACGGGGTCAATACCTCGGGAGATGAAGACACGTCCAGCTTCGTGTTCATAATGGGCGCAACGAACCGGCCCGATTTACTCGAACAATCTCTGCTGCGGCCCGGTAGACTCGATAAGCTCGTCTACGTGGGGCCCTACTGTGGGACGCGAGAGAAGACATCAGTGCTCGAAGCGCTTTGTCGGAAGTACAAACTAAGACCTGAAGTAAATTTGGAAGCCGTCGCAGAGGCACTACCCGACAAGTGCACCGGCGCTGACTTACTGCAGGTTGTCTCTACTGCACGGTCAGCGGCCGTGAGAGCACTCGTTGCTAAGCTACACAATGGAACGTTAAAGGAAAGCGAACTGAGTCCCGACTCCATCGTTATAGGGGTAACTGAACTATGGGAGGGCGTTGTATCATTCCGGCCCTCGGTCACAGACGATGAACTGTCGCATTACGAAGCGATGGAGTCACAAATCAACAGAGACGCCATAGAAAAGGAATTTCAGTTCAAAAATTTTAATGAAGCTTTTGGATTCATGACCAGGGTCGCGCTGCTAGCTGAGAAAATGGATCATCATCCTGAGTGGTTCAATGTATACAACAAGTTGCAAGTGACACTTTCTTCACATGACGTAAATGGATTGAGCAAAAGGGATATCAAAATGGCCAGTTTTATGGATAAAAATTATAATTAA
Protein
MCITTLRVTQLTLGENIHATKPQDKYFKPQGFDDNLHMLELCPTGLKHIFNQIQETISPFLNEDLSDLSLFLSSPIIPMLALTGCLGSGRHLAVKTLAKFNGINCLSIDCNALQSSTAKQTESKINATIQKAKAAAPVIVLLDNFEVFAIDAENNEDCRVIEYFMNTITDLYQNYTKHPIIFIAVTKDIEFKPNISRLFLEKFHIPKLNLRQRYEMLDWFSNVMRLNIDGAEHVKANELLVKQTPVNVNSQNVLQRVAAKTETFLYGDLDTLVHFATRESYLRQHNNYNQLPPDPDLRLIKEEDFNSALESIRKLQSQHLDAPKIPKVYWDDIGGLEKLKKELLKTIQLPIKYPHLFKNASLKRSGILLYGPPGCGKTLVAKAVSTELNVSFLSVKGPELLNMYIGQSEENVRKVFESARASSPCIIFVDELDALAPRRGAAGDSAGASDRVVSQLLAELDGVNTSGDEDTSSFVFIMGATNRPDLLEQSLLRPGRLDKLVYVGPYCGTREKTSVLEALCRKYKLRPEVNLEAVAEALPDKCTGADLLQVVSTARSAAVRALVAKLHNGTLKESELSPDSIVIGVTELWEGVVSFRPSVTDDELSHYEAMESQINRDAIEKEFQFKNFNEAFGFMTRVALLAEKMDHHPEWFNVYNKLQVTLSSHDVNGLSKRDIKMASFMDKNYN
Summary
Similarity
Belongs to the AAA ATPase family.
Uniprot
A0A2H1WD00
A0A194PHP2
A0A194RLR3
A0A212FL30
A0A0L7L367
B4QAP6
+ More
A0A0J9TYL5 A0A0J9RA23 A0A2J7PH28 A0A0L0BNF3 B4HN18 B3NN35 A0A0Q5VPL1 Q9V5R2 A1Z8D1 B5DZX0 A0A1W4VQE8 B4H8H0 B4P7N0 A0A1W4VDE3 A0A2J7PH22 A0A0R1DNM7 B3MIL9 A0A1A9USQ0 A0A0P8ZS19 A0A1I8Q7G9 A0A1I8Q7F9 A0A3B0JK62 A0A3B0JQ14 A0A1B0G0P0 A0A1B0GMP6 A0A1I8MDQ1 A0A1I8MDQ4 A0A1I8MDQ2 T1PAA0 B4MKB9 A0A0A1XCA6 W8BMK9 A0A0A1WD77 A0A0A1X9U1 W8BZF4 B4LMS7 A0A182IZ93 A0A0K8VQ15 A0A1L8DTT4 A0A0K8VUJ9 A0A0K8V620 B4KNA1 A0A0Q9XJF9 A0A084W3G0 B4J6S5 Q17NT9 A0A1B0GLR1 A0A0M3QVP5 A0A1L8DU68 A0A1Y1NDG1 A0A182HFD2 A0A182VZB5 A0A182SK23 A0A182V008 A0A182R627 A0A182L9Q6 A0A182U626 A0A182HLC4 Q7PXV3 A0A0C9QTY8 A0A182JU06 D6WES7 A0A2A3EQ48 A0A0L7RBM6 A0A2M4AMJ0 A0A182QM46 A0A154PLX6 W5J5E6 B0WIY8 N6TLX8 K7J5W7 A0A182WSH2 A0A182PGP4 A0A182NS49 A0A182FBP3 E2A7C3 A0A182YAK5 A0A232FFX6 A0A0P6DT56 E2C8G4 A0A0P5QSH4 A0A0P6JI49 A0A0P5WJ85 A0A0M9A1U2 A0A0N8CDB5 A0A164REC3 F4WPZ4 A0A158NIK0 A0A0P6BWQ3 K1PWD4 A0A0P5WRG3 W5NEY3 A0A195BCW3
A0A0J9TYL5 A0A0J9RA23 A0A2J7PH28 A0A0L0BNF3 B4HN18 B3NN35 A0A0Q5VPL1 Q9V5R2 A1Z8D1 B5DZX0 A0A1W4VQE8 B4H8H0 B4P7N0 A0A1W4VDE3 A0A2J7PH22 A0A0R1DNM7 B3MIL9 A0A1A9USQ0 A0A0P8ZS19 A0A1I8Q7G9 A0A1I8Q7F9 A0A3B0JK62 A0A3B0JQ14 A0A1B0G0P0 A0A1B0GMP6 A0A1I8MDQ1 A0A1I8MDQ4 A0A1I8MDQ2 T1PAA0 B4MKB9 A0A0A1XCA6 W8BMK9 A0A0A1WD77 A0A0A1X9U1 W8BZF4 B4LMS7 A0A182IZ93 A0A0K8VQ15 A0A1L8DTT4 A0A0K8VUJ9 A0A0K8V620 B4KNA1 A0A0Q9XJF9 A0A084W3G0 B4J6S5 Q17NT9 A0A1B0GLR1 A0A0M3QVP5 A0A1L8DU68 A0A1Y1NDG1 A0A182HFD2 A0A182VZB5 A0A182SK23 A0A182V008 A0A182R627 A0A182L9Q6 A0A182U626 A0A182HLC4 Q7PXV3 A0A0C9QTY8 A0A182JU06 D6WES7 A0A2A3EQ48 A0A0L7RBM6 A0A2M4AMJ0 A0A182QM46 A0A154PLX6 W5J5E6 B0WIY8 N6TLX8 K7J5W7 A0A182WSH2 A0A182PGP4 A0A182NS49 A0A182FBP3 E2A7C3 A0A182YAK5 A0A232FFX6 A0A0P6DT56 E2C8G4 A0A0P5QSH4 A0A0P6JI49 A0A0P5WJ85 A0A0M9A1U2 A0A0N8CDB5 A0A164REC3 F4WPZ4 A0A158NIK0 A0A0P6BWQ3 K1PWD4 A0A0P5WRG3 W5NEY3 A0A195BCW3
Pubmed
26354079
22118469
26227816
17994087
22936249
26108605
+ More
10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 17550304 25315136 25830018 24495485 24438588 17510324 28004739 26483478 20966253 12364791 14747013 17210077 18362917 19820115 20920257 23761445 23537049 20075255 20798317 25244985 28648823 21719571 21347285 22992520
10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 17550304 25315136 25830018 24495485 24438588 17510324 28004739 26483478 20966253 12364791 14747013 17210077 18362917 19820115 20920257 23761445 23537049 20075255 20798317 25244985 28648823 21719571 21347285 22992520
EMBL
ODYU01007835
SOQ50948.1
KQ459604
KPI92239.1
KQ460045
KPJ18255.1
+ More
AGBW02007949 OWR54399.1 JTDY01003307 KOB69766.1 CM000362 EDX06551.1 CM002911 KMY92880.1 KMY92881.1 KMY92882.1 NEVH01025150 PNF15634.1 JRES01001601 KNC21562.1 CH480816 EDW47322.1 CH954179 EDV56555.1 KQS62900.1 AY058375 AE013599 AAL13604.1 ACZ94389.1 AAF58736.4 CM000071 EDY69249.1 CH479223 EDW35005.1 CM000158 EDW90065.1 PNF15633.1 KRJ98885.1 CH902619 EDV38095.1 KPU77274.1 OUUW01000001 SPP75750.1 SPP75749.1 CCAG010023102 AJVK01012215 AJVK01012216 KA645619 AFP60248.1 CH963846 EDW72558.1 GBXI01005243 JAD09049.1 GAMC01011943 GAMC01011939 GAMC01011938 JAB94616.1 GBXI01017495 JAC96796.1 GBXI01006223 GBXI01003716 JAD08069.1 JAD10576.1 GAMC01011941 GAMC01011935 JAB94614.1 CH940648 EDW61018.2 GDHF01011335 JAI40979.1 GFDF01004201 JAV09883.1 GDHF01031927 GDHF01030711 GDHF01009755 JAI20387.1 JAI21603.1 JAI42559.1 GDHF01018008 JAI34306.1 CH933808 EDW09954.1 KRG05006.1 ATLV01019994 KE525289 KFB44754.1 CH916367 EDW00978.1 CH477196 EAT48385.1 AJWK01015551 AJWK01015552 CP012524 ALC42753.1 GFDF01004117 JAV09967.1 GEZM01009837 JAV94276.1 JXUM01037600 JXUM01037601 JXUM01037602 KQ561134 KXJ79611.1 APCN01000890 AAAB01008987 EAA00892.4 GBYB01007209 GBYB01008900 JAG76976.1 JAG78667.1 KQ971326 EEZ99865.1 KZ288203 PBC33392.1 KQ414617 KOC68156.1 GGFK01008682 MBW42003.1 AXCN02000266 KQ434977 KZC12881.1 ADMH02002183 ETN57979.1 DS231954 EDS28849.1 APGK01030336 KB740770 ENN78988.1 GL437329 EFN70620.1 NNAY01000305 OXU29339.1 GDIQ01072746 JAN21991.1 GL453666 EFN75694.1 GDIQ01110532 JAL41194.1 GDIQ01007478 JAN87259.1 GDIP01098377 JAM05338.1 KQ435793 KOX74203.1 GDIP01142927 JAL60787.1 LRGB01002190 KZS08580.1 GL888261 EGI63723.1 ADTU01016871 GDIP01008481 JAM95234.1 JH818807 EKC26023.1 GDIP01083289 JAM20426.1 AHAT01027972 KQ976513 KYM82398.1
AGBW02007949 OWR54399.1 JTDY01003307 KOB69766.1 CM000362 EDX06551.1 CM002911 KMY92880.1 KMY92881.1 KMY92882.1 NEVH01025150 PNF15634.1 JRES01001601 KNC21562.1 CH480816 EDW47322.1 CH954179 EDV56555.1 KQS62900.1 AY058375 AE013599 AAL13604.1 ACZ94389.1 AAF58736.4 CM000071 EDY69249.1 CH479223 EDW35005.1 CM000158 EDW90065.1 PNF15633.1 KRJ98885.1 CH902619 EDV38095.1 KPU77274.1 OUUW01000001 SPP75750.1 SPP75749.1 CCAG010023102 AJVK01012215 AJVK01012216 KA645619 AFP60248.1 CH963846 EDW72558.1 GBXI01005243 JAD09049.1 GAMC01011943 GAMC01011939 GAMC01011938 JAB94616.1 GBXI01017495 JAC96796.1 GBXI01006223 GBXI01003716 JAD08069.1 JAD10576.1 GAMC01011941 GAMC01011935 JAB94614.1 CH940648 EDW61018.2 GDHF01011335 JAI40979.1 GFDF01004201 JAV09883.1 GDHF01031927 GDHF01030711 GDHF01009755 JAI20387.1 JAI21603.1 JAI42559.1 GDHF01018008 JAI34306.1 CH933808 EDW09954.1 KRG05006.1 ATLV01019994 KE525289 KFB44754.1 CH916367 EDW00978.1 CH477196 EAT48385.1 AJWK01015551 AJWK01015552 CP012524 ALC42753.1 GFDF01004117 JAV09967.1 GEZM01009837 JAV94276.1 JXUM01037600 JXUM01037601 JXUM01037602 KQ561134 KXJ79611.1 APCN01000890 AAAB01008987 EAA00892.4 GBYB01007209 GBYB01008900 JAG76976.1 JAG78667.1 KQ971326 EEZ99865.1 KZ288203 PBC33392.1 KQ414617 KOC68156.1 GGFK01008682 MBW42003.1 AXCN02000266 KQ434977 KZC12881.1 ADMH02002183 ETN57979.1 DS231954 EDS28849.1 APGK01030336 KB740770 ENN78988.1 GL437329 EFN70620.1 NNAY01000305 OXU29339.1 GDIQ01072746 JAN21991.1 GL453666 EFN75694.1 GDIQ01110532 JAL41194.1 GDIQ01007478 JAN87259.1 GDIP01098377 JAM05338.1 KQ435793 KOX74203.1 GDIP01142927 JAL60787.1 LRGB01002190 KZS08580.1 GL888261 EGI63723.1 ADTU01016871 GDIP01008481 JAM95234.1 JH818807 EKC26023.1 GDIP01083289 JAM20426.1 AHAT01027972 KQ976513 KYM82398.1
Proteomes
UP000053268
UP000053240
UP000007151
UP000037510
UP000000304
UP000235965
+ More
UP000037069 UP000001292 UP000008711 UP000000803 UP000001819 UP000192221 UP000008744 UP000002282 UP000007801 UP000078200 UP000095300 UP000268350 UP000092444 UP000092462 UP000095301 UP000007798 UP000008792 UP000075880 UP000009192 UP000030765 UP000001070 UP000008820 UP000092461 UP000092553 UP000069940 UP000249989 UP000075920 UP000075901 UP000075903 UP000075882 UP000075902 UP000075840 UP000007062 UP000075881 UP000007266 UP000242457 UP000053825 UP000075886 UP000076502 UP000000673 UP000002320 UP000019118 UP000002358 UP000076407 UP000075885 UP000075884 UP000069272 UP000000311 UP000076408 UP000215335 UP000008237 UP000053105 UP000076858 UP000007755 UP000005205 UP000005408 UP000018468 UP000078540
UP000037069 UP000001292 UP000008711 UP000000803 UP000001819 UP000192221 UP000008744 UP000002282 UP000007801 UP000078200 UP000095300 UP000268350 UP000092444 UP000092462 UP000095301 UP000007798 UP000008792 UP000075880 UP000009192 UP000030765 UP000001070 UP000008820 UP000092461 UP000092553 UP000069940 UP000249989 UP000075920 UP000075901 UP000075903 UP000075882 UP000075902 UP000075840 UP000007062 UP000075881 UP000007266 UP000242457 UP000053825 UP000075886 UP000076502 UP000000673 UP000002320 UP000019118 UP000002358 UP000076407 UP000075885 UP000075884 UP000069272 UP000000311 UP000076408 UP000215335 UP000008237 UP000053105 UP000076858 UP000007755 UP000005205 UP000005408 UP000018468 UP000078540
PRIDE
Pfam
Interpro
IPR003593
AAA+_ATPase
+ More
IPR004977 Ribosomal_S25
IPR003959 ATPase_AAA_core
IPR027417 P-loop_NTPase
IPR003960 ATPase_AAA_CS
IPR041569 AAA_lid_3
IPR020568 Ribosomal_S5_D2-typ_fold
IPR029765 Mev_diP_decarb
IPR041431 Mvd1_C
IPR036554 GHMP_kinase_C_sf
IPR006204 GHMP_kinase_N_dom
IPR014721 Ribosomal_S5_D2-typ_fold_subgr
IPR015415 Vps4_C
IPR004977 Ribosomal_S25
IPR003959 ATPase_AAA_core
IPR027417 P-loop_NTPase
IPR003960 ATPase_AAA_CS
IPR041569 AAA_lid_3
IPR020568 Ribosomal_S5_D2-typ_fold
IPR029765 Mev_diP_decarb
IPR041431 Mvd1_C
IPR036554 GHMP_kinase_C_sf
IPR006204 GHMP_kinase_N_dom
IPR014721 Ribosomal_S5_D2-typ_fold_subgr
IPR015415 Vps4_C
Gene 3D
ProteinModelPortal
A0A2H1WD00
A0A194PHP2
A0A194RLR3
A0A212FL30
A0A0L7L367
B4QAP6
+ More
A0A0J9TYL5 A0A0J9RA23 A0A2J7PH28 A0A0L0BNF3 B4HN18 B3NN35 A0A0Q5VPL1 Q9V5R2 A1Z8D1 B5DZX0 A0A1W4VQE8 B4H8H0 B4P7N0 A0A1W4VDE3 A0A2J7PH22 A0A0R1DNM7 B3MIL9 A0A1A9USQ0 A0A0P8ZS19 A0A1I8Q7G9 A0A1I8Q7F9 A0A3B0JK62 A0A3B0JQ14 A0A1B0G0P0 A0A1B0GMP6 A0A1I8MDQ1 A0A1I8MDQ4 A0A1I8MDQ2 T1PAA0 B4MKB9 A0A0A1XCA6 W8BMK9 A0A0A1WD77 A0A0A1X9U1 W8BZF4 B4LMS7 A0A182IZ93 A0A0K8VQ15 A0A1L8DTT4 A0A0K8VUJ9 A0A0K8V620 B4KNA1 A0A0Q9XJF9 A0A084W3G0 B4J6S5 Q17NT9 A0A1B0GLR1 A0A0M3QVP5 A0A1L8DU68 A0A1Y1NDG1 A0A182HFD2 A0A182VZB5 A0A182SK23 A0A182V008 A0A182R627 A0A182L9Q6 A0A182U626 A0A182HLC4 Q7PXV3 A0A0C9QTY8 A0A182JU06 D6WES7 A0A2A3EQ48 A0A0L7RBM6 A0A2M4AMJ0 A0A182QM46 A0A154PLX6 W5J5E6 B0WIY8 N6TLX8 K7J5W7 A0A182WSH2 A0A182PGP4 A0A182NS49 A0A182FBP3 E2A7C3 A0A182YAK5 A0A232FFX6 A0A0P6DT56 E2C8G4 A0A0P5QSH4 A0A0P6JI49 A0A0P5WJ85 A0A0M9A1U2 A0A0N8CDB5 A0A164REC3 F4WPZ4 A0A158NIK0 A0A0P6BWQ3 K1PWD4 A0A0P5WRG3 W5NEY3 A0A195BCW3
A0A0J9TYL5 A0A0J9RA23 A0A2J7PH28 A0A0L0BNF3 B4HN18 B3NN35 A0A0Q5VPL1 Q9V5R2 A1Z8D1 B5DZX0 A0A1W4VQE8 B4H8H0 B4P7N0 A0A1W4VDE3 A0A2J7PH22 A0A0R1DNM7 B3MIL9 A0A1A9USQ0 A0A0P8ZS19 A0A1I8Q7G9 A0A1I8Q7F9 A0A3B0JK62 A0A3B0JQ14 A0A1B0G0P0 A0A1B0GMP6 A0A1I8MDQ1 A0A1I8MDQ4 A0A1I8MDQ2 T1PAA0 B4MKB9 A0A0A1XCA6 W8BMK9 A0A0A1WD77 A0A0A1X9U1 W8BZF4 B4LMS7 A0A182IZ93 A0A0K8VQ15 A0A1L8DTT4 A0A0K8VUJ9 A0A0K8V620 B4KNA1 A0A0Q9XJF9 A0A084W3G0 B4J6S5 Q17NT9 A0A1B0GLR1 A0A0M3QVP5 A0A1L8DU68 A0A1Y1NDG1 A0A182HFD2 A0A182VZB5 A0A182SK23 A0A182V008 A0A182R627 A0A182L9Q6 A0A182U626 A0A182HLC4 Q7PXV3 A0A0C9QTY8 A0A182JU06 D6WES7 A0A2A3EQ48 A0A0L7RBM6 A0A2M4AMJ0 A0A182QM46 A0A154PLX6 W5J5E6 B0WIY8 N6TLX8 K7J5W7 A0A182WSH2 A0A182PGP4 A0A182NS49 A0A182FBP3 E2A7C3 A0A182YAK5 A0A232FFX6 A0A0P6DT56 E2C8G4 A0A0P5QSH4 A0A0P6JI49 A0A0P5WJ85 A0A0M9A1U2 A0A0N8CDB5 A0A164REC3 F4WPZ4 A0A158NIK0 A0A0P6BWQ3 K1PWD4 A0A0P5WRG3 W5NEY3 A0A195BCW3
PDB
5VCA
E-value=5.82674e-56,
Score=553
Ontologies
PATHWAY
GO
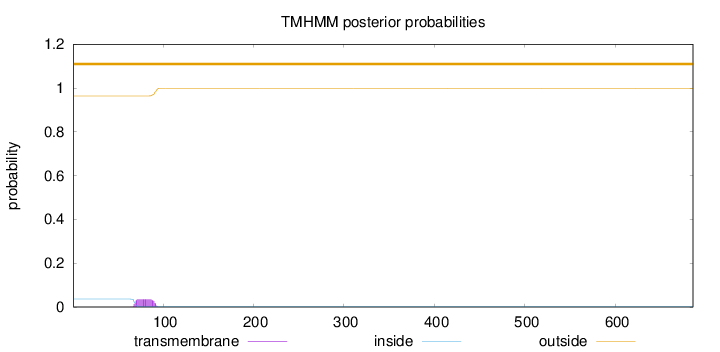
Topology
Length:
686
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.790629999999998
Exp number, first 60 AAs:
0.00013
Total prob of N-in:
0.03635
outside
1 - 686
Population Genetic Test Statistics
Pi
262.134966
Theta
186.325877
Tajima's D
2.484172
CLR
0.327259
CSRT
0.942802859857007
Interpretation
Uncertain
