Gene
KWMTBOMO15111
Pre Gene Modal
BGIBMGA004904
Annotation
isopentenyl-diphosphate_delta_isomerase_[Bombyx_mori]
Full name
rRNA adenine N(6)-methyltransferase
Location in the cell
Cytoplasmic Reliability : 2.479
Sequence
CDS
ATGTTGGTCAGAAAGCTAACAAGATCACTGTGGAGCACCCTAAGAGTCGAGAAAAGATTTTTGGCTTCAGAACCTGTGAAACCTGCTACTCTAGATAAGGATGGTCTGGAATTAGCCCAGGCAGAAGCTTTAGATAAAGATATTTGCCTGCTAGTTGATGAAAAGGACAACTTCATAGGAACGGCTACAAAGAGAGAATGTCATAAAGTAGGACCTGATGGTGATGTGCTACTCCATAGAGCATTCAGTGTTTTCCTCTTCAACAAGAGAGGTGATATGTTCCTTCAGAGGAGATCAAGCCAAAAAGTAACATATCCCGATTACTACACAAATGCTTGCTGCAGCCATCCACTGTACATAGATGAGAAGCCCGAGGAAATCATAACGGCAGCCAGACGTAGGATGAACCATGAACTGGGAATCCCACTAGATCAGTTGGACCCGGAACTGTTTACGTTCATGACCCGCGTGCACTATCACGACCCGGGTGATGGAGTGTGGGGCGAACACGAGATCGACCACATACTGTTCTTCCAAAGCGACGTTAAGGTCAAGCCGAACTCGGACGAGATCTCGGAGTATTGTTTCGTGCCCAAAGCAGAGTTTAATTCGTTCCTTCCAACCTTGGAGGGTCCGATAACGCCGTGGTTCAACATGATCCGCCGCCACCGATTGAAACTCTGGTGGGACAACTTGCACCGCATCAAAGAGCTCGCCGAACCGGAGAAAATACAAAAATTTGTTGATAAAAAATAA
Protein
MLVRKLTRSLWSTLRVEKRFLASEPVKPATLDKDGLELAQAEALDKDICLLVDEKDNFIGTATKRECHKVGPDGDVLLHRAFSVFLFNKRGDMFLQRRSSQKVTYPDYYTNACCSHPLYIDEKPEEIITAARRRMNHELGIPLDQLDPELFTFMTRVHYHDPGDGVWGEHEIDHILFFQSDVKVKPNSDEISEYCFVPKAEFNSFLPTLEGPITPWFNMIRRHRLKLWWDNLHRIKELAEPEKIQKFVDKK
Summary
Similarity
Belongs to the class I-like SAM-binding methyltransferase superfamily. rRNA adenine N(6)-methyltransferase family.
Uniprot
H9J5W4
Q2F5Q7
A0A0C5KR39
S4Q0B4
A0A194PG15
I4DJR9
+ More
A0A194RQF3 A0A212FKY2 A0A2H1WCZ9 A0A0L7L280 B0XFL6 A0A182QBL0 U5EZ74 A0A2M4ASB9 B4KCU8 A0A182Y3R8 A0A182F7H7 B4MBG8 Q1HQH4 W5JIS4 A0A023EKX7 A0A1Q3F596 A0A1Q3F4G3 A0A0H4IVW7 B4JS40 A0A0M4ERR4 A0A182VZK9 A0A182JRZ8 A0A182UJ31 A0A182WSS1 J3JWE5 B4NKG8 A0A2M4C0K0 A0A182RIM6 A0A182VDA4 I1VJ70 Q7PY18 A0A182HLM4 A0APV9 A0A182NRU9 Q8MRH1 B4R121 B4IKH9 A0A3B0K1T9 B3P033 A0A0K8TM64 Q9VDC2 A0APV5 Q29B04 B4GZ93 A0A1W4VLA3 B4PQD6 B5RIJ6 A0A1Y1MG54 Q56CY5 A0A1Y1MGK6 A0A0N9H5S7 A0A182JG31 D6WP58 A0A1A9UJT2 A0A084VD35 A0A034WJ79 B3LZ32 A0A0L0CGZ6 A0A0K8UMY1 A0A1B0G652 A0A1B0D3X8 A0A1B6LRP7 A0A1A9YLP3 A0A1B0BYC7 A0A1B0CQ36 A0A1L8E526 W8AL09 U4U6N5 A0A1A9Z9J0 A0A2J7PEJ4 A0A1A9WY08 A0A0A1WGH7 A0A2P8YTN1 A0A087ZZX6 A0A1B6C3P3 A4UU28 A0A023FBL3 A0A2A3E5A3 A0A1W4XFU4 A0A1B6ICL1 A0A069DQV6 A0A1L8E516 V9IG77 A0A1I8MJW3 A0A2R7WF69 A0A067QH23 A0A1J1IR23 A0A1I8PFF5 A0A0A9Y2Z6 A0A0L7QU11 A0A026W5A6 R4WQC4 K7J1J3
A0A194RQF3 A0A212FKY2 A0A2H1WCZ9 A0A0L7L280 B0XFL6 A0A182QBL0 U5EZ74 A0A2M4ASB9 B4KCU8 A0A182Y3R8 A0A182F7H7 B4MBG8 Q1HQH4 W5JIS4 A0A023EKX7 A0A1Q3F596 A0A1Q3F4G3 A0A0H4IVW7 B4JS40 A0A0M4ERR4 A0A182VZK9 A0A182JRZ8 A0A182UJ31 A0A182WSS1 J3JWE5 B4NKG8 A0A2M4C0K0 A0A182RIM6 A0A182VDA4 I1VJ70 Q7PY18 A0A182HLM4 A0APV9 A0A182NRU9 Q8MRH1 B4R121 B4IKH9 A0A3B0K1T9 B3P033 A0A0K8TM64 Q9VDC2 A0APV5 Q29B04 B4GZ93 A0A1W4VLA3 B4PQD6 B5RIJ6 A0A1Y1MG54 Q56CY5 A0A1Y1MGK6 A0A0N9H5S7 A0A182JG31 D6WP58 A0A1A9UJT2 A0A084VD35 A0A034WJ79 B3LZ32 A0A0L0CGZ6 A0A0K8UMY1 A0A1B0G652 A0A1B0D3X8 A0A1B6LRP7 A0A1A9YLP3 A0A1B0BYC7 A0A1B0CQ36 A0A1L8E526 W8AL09 U4U6N5 A0A1A9Z9J0 A0A2J7PEJ4 A0A1A9WY08 A0A0A1WGH7 A0A2P8YTN1 A0A087ZZX6 A0A1B6C3P3 A4UU28 A0A023FBL3 A0A2A3E5A3 A0A1W4XFU4 A0A1B6ICL1 A0A069DQV6 A0A1L8E516 V9IG77 A0A1I8MJW3 A0A2R7WF69 A0A067QH23 A0A1J1IR23 A0A1I8PFF5 A0A0A9Y2Z6 A0A0L7QU11 A0A026W5A6 R4WQC4 K7J1J3
EC Number
2.1.1.-
Pubmed
19121390
23622113
26354079
22651552
22118469
26227816
+ More
17994087 18057021 25244985 17204158 17510324 20920257 23761445 24945155 26483478 26899871 22516182 23537049 12364791 14747013 17210077 16951084 26369729 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 19126864 26109357 26109356 15632085 17550304 28004739 18362917 19820115 24438588 25348373 26108605 24495485 25830018 29403074 17634749 25474469 26334808 25315136 24845553 25401762 26823975 24508170 30249741 23691247 20075255
17994087 18057021 25244985 17204158 17510324 20920257 23761445 24945155 26483478 26899871 22516182 23537049 12364791 14747013 17210077 16951084 26369729 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 19126864 26109357 26109356 15632085 17550304 28004739 18362917 19820115 24438588 25348373 26108605 24495485 25830018 29403074 17634749 25474469 26334808 25315136 24845553 25401762 26823975 24508170 30249741 23691247 20075255
EMBL
BABH01030020
DQ311366
ABD36310.1
KJ951988
AJP70490.1
GAIX01000083
+ More
JAA92477.1 KQ459604 KPI92237.1 AK401537 BAM18159.1 KQ460045 KPJ18256.1 AGBW02007949 OWR54401.1 ODYU01007835 SOQ50950.1 JTDY01003409 KOB69603.1 DS232938 EDS26879.1 AXCN02000250 GANO01001306 JAB58565.1 GGFK01010350 MBW43671.1 CH933806 EDW16970.1 KRG02420.1 CH940656 EDW58439.1 DQ440470 CH477376 ABF18503.1 EAT42319.1 ADMH02001371 ETN62790.1 JXUM01079199 JXUM01079200 JXUM01079201 GAPW01003747 KQ563113 JAC09851.1 KXJ74444.1 GFDL01012313 JAV22732.1 GFDL01012589 JAV22456.1 KP689341 AKO63323.1 CH916373 EDV94580.1 CP012526 ALC47901.1 APGK01025935 BT127563 KB740605 AEE62525.1 ENN80064.1 CH964272 EDW85140.1 GGFJ01009377 MBW58518.1 JQ855691 AFI45054.1 AAAB01008987 EAA00859.4 APCN01000909 AM294697 CAL26683.1 AY119630 AAM50284.1 CM000364 EDX12108.1 CH480853 EDW51583.1 OUUW01000013 SPP88227.1 CH954181 EDV48269.1 GDAI01002382 JAI15221.1 AE014297 AY118469 AM294686 AM294687 AM294688 AM294689 AM294690 AM294691 AM294692 AM294694 AM294695 AM294696 FM245625 FM245626 FM245627 FM245628 FM245629 FM245630 FM245631 FM245632 FM245633 FM245634 AAF55875.1 AAM49838.1 CAL26672.1 CAL26673.1 CAL26674.1 CAL26675.1 CAL26676.1 CAL26677.1 CAL26678.1 CAL26680.1 CAL26681.1 CAL26682.1 CAR93551.1 CAR93552.1 CAR93553.1 CAR93554.1 CAR93555.1 CAR93556.1 CAR93557.1 CAR93558.1 CAR93559.1 CAR93560.1 AM294693 CAL26679.1 CM000070 EAL27195.1 CH479198 EDW28111.1 CM000160 EDW96245.1 BT044120 ACH92185.1 GEZM01032178 JAV84792.1 AY966011 AAX78437.1 GEZM01032177 JAV84793.1 KT784804 ALF95236.1 KQ971352 EFA07280.1 ATLV01011143 KE524645 KFB35879.1 GAKP01004540 GAKP01004538 JAC54412.1 CH902617 EDV41906.2 JRES01000409 KNC31515.1 GDHF01029120 GDHF01024255 GDHF01014571 JAI23194.1 JAI28059.1 JAI37743.1 CCAG010005914 AJVK01011091 GEBQ01023543 GEBQ01013748 JAT16434.1 JAT26229.1 JXJN01022585 AJWK01022977 GFDF01000452 JAV13632.1 GAMC01017185 GAMC01017183 JAB89370.1 KB632095 ERL88732.1 NEVH01026094 PNF14763.1 GBXI01016310 JAC97981.1 PYGN01000366 PSN47596.1 GEDC01029443 JAS07855.1 EF489295 ABP01599.1 GBBI01000323 JAC18389.1 KZ288387 PBC26436.1 GECU01023093 JAS84613.1 GBGD01002862 JAC86027.1 GFDF01000462 JAV13622.1 JR045932 AEY60088.1 KK854722 PTY18169.1 KK853419 KDR07729.1 CVRI01000055 CRL00937.1 GBHO01017085 GBRD01011449 GDHC01002841 JAG26519.1 JAG54375.1 JAQ15788.1 KQ414740 KOC62044.1 KK107468 QOIP01000001 EZA50214.1 RLU27474.1 AK417896 BAN21111.1
JAA92477.1 KQ459604 KPI92237.1 AK401537 BAM18159.1 KQ460045 KPJ18256.1 AGBW02007949 OWR54401.1 ODYU01007835 SOQ50950.1 JTDY01003409 KOB69603.1 DS232938 EDS26879.1 AXCN02000250 GANO01001306 JAB58565.1 GGFK01010350 MBW43671.1 CH933806 EDW16970.1 KRG02420.1 CH940656 EDW58439.1 DQ440470 CH477376 ABF18503.1 EAT42319.1 ADMH02001371 ETN62790.1 JXUM01079199 JXUM01079200 JXUM01079201 GAPW01003747 KQ563113 JAC09851.1 KXJ74444.1 GFDL01012313 JAV22732.1 GFDL01012589 JAV22456.1 KP689341 AKO63323.1 CH916373 EDV94580.1 CP012526 ALC47901.1 APGK01025935 BT127563 KB740605 AEE62525.1 ENN80064.1 CH964272 EDW85140.1 GGFJ01009377 MBW58518.1 JQ855691 AFI45054.1 AAAB01008987 EAA00859.4 APCN01000909 AM294697 CAL26683.1 AY119630 AAM50284.1 CM000364 EDX12108.1 CH480853 EDW51583.1 OUUW01000013 SPP88227.1 CH954181 EDV48269.1 GDAI01002382 JAI15221.1 AE014297 AY118469 AM294686 AM294687 AM294688 AM294689 AM294690 AM294691 AM294692 AM294694 AM294695 AM294696 FM245625 FM245626 FM245627 FM245628 FM245629 FM245630 FM245631 FM245632 FM245633 FM245634 AAF55875.1 AAM49838.1 CAL26672.1 CAL26673.1 CAL26674.1 CAL26675.1 CAL26676.1 CAL26677.1 CAL26678.1 CAL26680.1 CAL26681.1 CAL26682.1 CAR93551.1 CAR93552.1 CAR93553.1 CAR93554.1 CAR93555.1 CAR93556.1 CAR93557.1 CAR93558.1 CAR93559.1 CAR93560.1 AM294693 CAL26679.1 CM000070 EAL27195.1 CH479198 EDW28111.1 CM000160 EDW96245.1 BT044120 ACH92185.1 GEZM01032178 JAV84792.1 AY966011 AAX78437.1 GEZM01032177 JAV84793.1 KT784804 ALF95236.1 KQ971352 EFA07280.1 ATLV01011143 KE524645 KFB35879.1 GAKP01004540 GAKP01004538 JAC54412.1 CH902617 EDV41906.2 JRES01000409 KNC31515.1 GDHF01029120 GDHF01024255 GDHF01014571 JAI23194.1 JAI28059.1 JAI37743.1 CCAG010005914 AJVK01011091 GEBQ01023543 GEBQ01013748 JAT16434.1 JAT26229.1 JXJN01022585 AJWK01022977 GFDF01000452 JAV13632.1 GAMC01017185 GAMC01017183 JAB89370.1 KB632095 ERL88732.1 NEVH01026094 PNF14763.1 GBXI01016310 JAC97981.1 PYGN01000366 PSN47596.1 GEDC01029443 JAS07855.1 EF489295 ABP01599.1 GBBI01000323 JAC18389.1 KZ288387 PBC26436.1 GECU01023093 JAS84613.1 GBGD01002862 JAC86027.1 GFDF01000462 JAV13622.1 JR045932 AEY60088.1 KK854722 PTY18169.1 KK853419 KDR07729.1 CVRI01000055 CRL00937.1 GBHO01017085 GBRD01011449 GDHC01002841 JAG26519.1 JAG54375.1 JAQ15788.1 KQ414740 KOC62044.1 KK107468 QOIP01000001 EZA50214.1 RLU27474.1 AK417896 BAN21111.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000007151
UP000037510
UP000002320
+ More
UP000075886 UP000009192 UP000076408 UP000069272 UP000008792 UP000008820 UP000000673 UP000069940 UP000249989 UP000001070 UP000092553 UP000075920 UP000075881 UP000075902 UP000076407 UP000019118 UP000007798 UP000075900 UP000075903 UP000007062 UP000075840 UP000075884 UP000000304 UP000001292 UP000268350 UP000008711 UP000000803 UP000001819 UP000008744 UP000192221 UP000002282 UP000075880 UP000007266 UP000078200 UP000030765 UP000007801 UP000037069 UP000092444 UP000092462 UP000092443 UP000092460 UP000092461 UP000030742 UP000092445 UP000235965 UP000091820 UP000245037 UP000005203 UP000242457 UP000192223 UP000095301 UP000027135 UP000183832 UP000095300 UP000053825 UP000053097 UP000279307 UP000002358
UP000075886 UP000009192 UP000076408 UP000069272 UP000008792 UP000008820 UP000000673 UP000069940 UP000249989 UP000001070 UP000092553 UP000075920 UP000075881 UP000075902 UP000076407 UP000019118 UP000007798 UP000075900 UP000075903 UP000007062 UP000075840 UP000075884 UP000000304 UP000001292 UP000268350 UP000008711 UP000000803 UP000001819 UP000008744 UP000192221 UP000002282 UP000075880 UP000007266 UP000078200 UP000030765 UP000007801 UP000037069 UP000092444 UP000092462 UP000092443 UP000092460 UP000092461 UP000030742 UP000092445 UP000235965 UP000091820 UP000245037 UP000005203 UP000242457 UP000192223 UP000095301 UP000027135 UP000183832 UP000095300 UP000053825 UP000053097 UP000279307 UP000002358
Interpro
Gene 3D
ProteinModelPortal
H9J5W4
Q2F5Q7
A0A0C5KR39
S4Q0B4
A0A194PG15
I4DJR9
+ More
A0A194RQF3 A0A212FKY2 A0A2H1WCZ9 A0A0L7L280 B0XFL6 A0A182QBL0 U5EZ74 A0A2M4ASB9 B4KCU8 A0A182Y3R8 A0A182F7H7 B4MBG8 Q1HQH4 W5JIS4 A0A023EKX7 A0A1Q3F596 A0A1Q3F4G3 A0A0H4IVW7 B4JS40 A0A0M4ERR4 A0A182VZK9 A0A182JRZ8 A0A182UJ31 A0A182WSS1 J3JWE5 B4NKG8 A0A2M4C0K0 A0A182RIM6 A0A182VDA4 I1VJ70 Q7PY18 A0A182HLM4 A0APV9 A0A182NRU9 Q8MRH1 B4R121 B4IKH9 A0A3B0K1T9 B3P033 A0A0K8TM64 Q9VDC2 A0APV5 Q29B04 B4GZ93 A0A1W4VLA3 B4PQD6 B5RIJ6 A0A1Y1MG54 Q56CY5 A0A1Y1MGK6 A0A0N9H5S7 A0A182JG31 D6WP58 A0A1A9UJT2 A0A084VD35 A0A034WJ79 B3LZ32 A0A0L0CGZ6 A0A0K8UMY1 A0A1B0G652 A0A1B0D3X8 A0A1B6LRP7 A0A1A9YLP3 A0A1B0BYC7 A0A1B0CQ36 A0A1L8E526 W8AL09 U4U6N5 A0A1A9Z9J0 A0A2J7PEJ4 A0A1A9WY08 A0A0A1WGH7 A0A2P8YTN1 A0A087ZZX6 A0A1B6C3P3 A4UU28 A0A023FBL3 A0A2A3E5A3 A0A1W4XFU4 A0A1B6ICL1 A0A069DQV6 A0A1L8E516 V9IG77 A0A1I8MJW3 A0A2R7WF69 A0A067QH23 A0A1J1IR23 A0A1I8PFF5 A0A0A9Y2Z6 A0A0L7QU11 A0A026W5A6 R4WQC4 K7J1J3
A0A194RQF3 A0A212FKY2 A0A2H1WCZ9 A0A0L7L280 B0XFL6 A0A182QBL0 U5EZ74 A0A2M4ASB9 B4KCU8 A0A182Y3R8 A0A182F7H7 B4MBG8 Q1HQH4 W5JIS4 A0A023EKX7 A0A1Q3F596 A0A1Q3F4G3 A0A0H4IVW7 B4JS40 A0A0M4ERR4 A0A182VZK9 A0A182JRZ8 A0A182UJ31 A0A182WSS1 J3JWE5 B4NKG8 A0A2M4C0K0 A0A182RIM6 A0A182VDA4 I1VJ70 Q7PY18 A0A182HLM4 A0APV9 A0A182NRU9 Q8MRH1 B4R121 B4IKH9 A0A3B0K1T9 B3P033 A0A0K8TM64 Q9VDC2 A0APV5 Q29B04 B4GZ93 A0A1W4VLA3 B4PQD6 B5RIJ6 A0A1Y1MG54 Q56CY5 A0A1Y1MGK6 A0A0N9H5S7 A0A182JG31 D6WP58 A0A1A9UJT2 A0A084VD35 A0A034WJ79 B3LZ32 A0A0L0CGZ6 A0A0K8UMY1 A0A1B0G652 A0A1B0D3X8 A0A1B6LRP7 A0A1A9YLP3 A0A1B0BYC7 A0A1B0CQ36 A0A1L8E526 W8AL09 U4U6N5 A0A1A9Z9J0 A0A2J7PEJ4 A0A1A9WY08 A0A0A1WGH7 A0A2P8YTN1 A0A087ZZX6 A0A1B6C3P3 A4UU28 A0A023FBL3 A0A2A3E5A3 A0A1W4XFU4 A0A1B6ICL1 A0A069DQV6 A0A1L8E516 V9IG77 A0A1I8MJW3 A0A2R7WF69 A0A067QH23 A0A1J1IR23 A0A1I8PFF5 A0A0A9Y2Z6 A0A0L7QU11 A0A026W5A6 R4WQC4 K7J1J3
PDB
2ICK
E-value=5.71637e-47,
Score=471
Ontologies
PATHWAY
GO
PANTHER
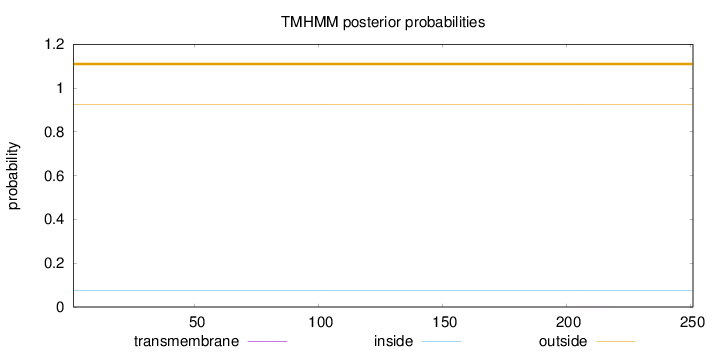
Topology
Length:
251
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00297
Exp number, first 60 AAs:
0
Total prob of N-in:
0.07370
outside
1 - 251
Population Genetic Test Statistics
Pi
155.086087
Theta
174.502961
Tajima's D
-0.683685
CLR
0.29179
CSRT
0.204139793010349
Interpretation
Uncertain
