Gene
KWMTBOMO15107 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA004902
Annotation
Transmembrane_protein_C9orf5_[Papilio_xuthus]
Location in the cell
PlasmaMembrane Reliability : 4.433
Sequence
CDS
ATGCAGCAGTCACCTTTTGAAAATATTTTCAATATCCTGGGTGGTATAACTGAAGGCAATGAAAAGCCTATGAAGCATGGGTTTTATAATGCTTTTGCCCTTTTCATACTCACAATATGTTGCGCCGCGGTATACGTTCTATTCCTAATACTAGAACCATTCTTCAAACCATTATTTTGGGCTCTATTAGTCGGTTCAATACTGCACCCTTTCAAATATAAAATTTCGAAGAAACTCAAGTCGTGGTTCGAAGATCTGGAGAAGTCTGACACGTCAGTGATAGTCGGTTTGATGGTGATCCCAATAAATGTTGTTAACTTCGCTTCAGACACAATTGGACAACAATTTAGAGAGCACTACAAGACTGTAATTACGCTACTCTCTGCAATATTTGTGTTGCCATGGTTGTATGATTCTATACCACGAGGTTTATCCTGTCTATTCTGGCAGTTGATCAGTTTCATTAGCTTTTCCACCAAAATGCTAATAAGCATCTGCACCTCATATATTACTCTAACCATAGTTTTGGCCTACATTATCAGCGTTTACACATTATGGACACCACAAAGGGAGAACATGTTCCGAGTGACGTCACAACTACTTTGGCTCGTAATTACGAGCTTTATGGCTAGTTTAACGGGTTCCATCCAAGTGTACACATTCATTTTTCTCCAAATTATATGTATAACTGGCTTTATAATGGAAATTCTGGATGTTCACAAGAAAATCCTGTTAAAGGATCCTGAAGCATCGATCATAATATCGATACACAGAGTTATAACAAACAGTCACAGAGAGAAAGAAACTAACACAGAAGACAGTAAATCTGATAGTGCGCAAATCTCTGCTAGCGAAACTGAAGGGACCCCGGATAAGGAGACCCCTTCTCCCGGTACACCAGAAACGTTACCCAAAAGCGGAAGCTGGACCAGCAATGAACCCAGCCAATGTCCTCCAAGACCACCGAAAATATTGTATAAAACGCGAACATTGTCCGCATTGCCACTGTCGGGTACAGGCAAAGTTAAACAAGAATATCGCTTCCTGCCATCAATTAAACACAAATCGCTCGATGATTATTACATGAAGAATAACTTCGACAATGACGAAGAAACAGCGTTGTATTTCAAATTGCTTTTCTTCGGTTTCCTCTGTATGTTGGTCTGGAAGAACATATGGTTACTGCCATTAGTTTTGGTGTTTTTAGTTATACACTTCATGAAGATTGGTTTGGATTATTTCGGAGTCTGGTTGTTCTTTGAAAACCAGTACAACAATGTGGCTTTGAAGATAAAAGCTTGGTGGAATAATAGGGAGACTGCTCTAATACCAGCACATATACGTGGAGTCGGTAAAATGATGTCAATATGCAATACAAGCCTCAGAGATATGGCATATAGCTCTGTAGATACTGTGTCCAGTTGCATTGTAATCATTGGCTTGATACTATTTCTTACAGTAGCAACCATTTTTATTTGTATACAGATATACTCGGAAGCCATAGTTGTGGTACAACTAAGCGGTAGTCTTCTTAACAGTACGTATGTTCAGAATAGTGTATTGCATAACTATCTGCCCGAGGGATGGGAGGAGAAACTCGACTCTCTTATCGATGACGCGTATACGTACGGAAGGGAAGGAATATCGACAACCGTTAAAAACATCTTGAAGGAAGGCGATCCAGAGAAAGTATCACGCGTCGAACAACAAGTTTTGGAGATTTGGGATAGAGTTTATCAGAGTTGGGCGTCGGGACATTTCGCCACGGCGGTCGGGCCCCAAGTCGATAGTACCGCCATACAGGACTCGTGGGATAACTTCGTCAACAACGTATCCGACTCATCGAGTTTGTTCGACTACTCCGGTGTAGTTTCGTGGTTACAAGCTAACGTGGGTACTCTGAGCGCTATAGCCAGTAGCATATGGGCGCCGCTGGCCAGCAATGTTTCGTTACTGGCCGGATCTTTGGGGGCATTTACGTCATTACTCCTCGGAGGAGGCAGCGCGATTATCAACATGTTCATAAATATGATAGTATTCTTCACGACACTGTTCTATCTGCTAGCGTCCAGTAACGCGCTCTACAAGCCAGTGGAAGTGATCACTCAGGTGCAGCCGAATTTCGGACCCCGTCTTGGCATTGCACTTTCTGTTGCTATCAACCAGGTATTCAGGGCATCGTTCAAAATGGCGTTGTTCTATGGATTATGGACTTGGCTAGTGCACAATCTCTTCGGGGCTAAAGTGGTATATTTACCTTCAGGTGTTAGAATAAACTTCACGAGTTGCCAAAACGCTGGCTGGTCTTGTTTCGCAACAAAGGACATTGTTTACAGTTGTGCTAATGTCAAAAGTAATGACTTCATAATACATCTAAAGGACTTTTACACGGATGATATAAAAAGTTTAACCGTGCAGAATTGTAAGGACGTCAAAGTGGTCCTCGACTGCCCTATATTACAGAAGGCATCTTCGCTTCAAAAGTTCAAAGTAAAAGACTGCGACAAACTCCAATTCATCTCCCTATCGAACAACTCCCCGGTTCAGACGCCGCCGGAAGTTACCATAGAGAACGTGAAGGAAGTGGTGTCTTTACCAAGGAAGTTCTTCAGATCACCGACCACTGCTAACGAAGTGAAATGCTCCGGTACCGCCTCCCTGAAAAGTATAAGTGTCGTCGATAGCTATATAAAATCAATAAACACAAGGGCCATTTACAACGTAACCGGAGTCAGTAGCGTTGAGTTCGTGAATGTGACTATCGGTGAGATTCAGAATCAAGGTATAGAGGCCATATTAGGGAACGAGGACACGTTGTTTTCGTTCGCCAACAACAAAATAGAAAGCTTGAGATATAAAGGCGTAGCTGTACAATGTACAAGTGCGTTCTTCACGGAGAACACATTTCGGGACGTCCTTAGTAATTCGTTGAATGTCACCGCGGACAATCTGAGTGTCGTTGGTAACAAATTCAAACAGGTCAACGGTCTGATACTGAAAGCTTCGAGCATAGAAATATCGAAAAACGAGTTCGGCACGCTCAAGAGCAACGCTTTCACGAACGTTAAGTGTTTAAGAAGAAATACCGGTAGAAGAGGTTTCACGTTCGCTCTGAATACTGTAGAAAACGTTGAACCACATTCGCTGGTCTTCGATTACGCTAGTTGCAAGACAGCCGGCGCCTCTGTTACGTATGAAAACAACAAAATCGATTGCAGATGCCGCGACATAGACTTCTTGGATGCTCCAACTGAACTCAACAGCTTAATTCTGGACTTGTCGTTTAACAACACGTGCTTATCGGCACCCTGTCTGCTTCCCGTCGAGATTGTCAAGCTGTTGTTGGAGAGGGGAATGTGCGATATAAATCTCGATCCTCAAATAATGTGCTTGTTGTACGGCGATAGGCATCTCGCGAACAACGAGTTGACGACAGACGAGGACGTCACCGAGCCCACGTCCACATTTTACATAATACGCCAGGCAGATCCCCTTGACGGCGGCGCTAGTTCGTTAACGGCTATAAACAAGGACGAACTCCTTAGGGACAGCCATTTCAACATAACGAATAGAACGTCTATTAGAGTCGTTTTCGATTCATCTAGAGACTTCGTTGAGACCTTGCGAGGCACGGATTCGTCTCGTAAAAAAACCACAGACAAAGTCAACGAGGAATACACGAACAGATGTGTCGGTCCTCAGTGCAGGAACAACGCGGTGCAGGACAAACAGAAGGCGCTTGATTTTTACAAATTCATTTATAGTCAGCTACGCCAGCCGAGGGCAAACGATAAGAAGAAGAAAAAGAGGGTTCTAGTGCCACAATTGGATGCCGTTCTAGCTGCAGTGCTAGGAGCTGCGCCGTTCCTAGGGCCATATTTGGCGGGTTTACCGGCAGCTCTTGATGTGTGGCTTCAGGGCAGACCCTTGGCGGCTGTGATGCTGCCCCTTGTTCAAGCCGCACCCATCGCATTTTTAGATGCGGCTGTTTACGCTGAGATTAAAGATGGTGGACATCCCTACGTGACAGGTCTAGCTATAGCGGGCGGTATCTTCTATCTCGGTCCCGAAGGAGCTATACTAGGTCCCTTGTTACTATGCTGTCTTATGGTGGTCCTGAATTTATCGTCAGCATTCCTCCGAGACACGCCAACGGACGAGAGGGCCGCCTTACATTCTCGTGTCAGATATGGTGCCTATATGTATTGA
Protein
MQQSPFENIFNILGGITEGNEKPMKHGFYNAFALFILTICCAAVYVLFLILEPFFKPLFWALLVGSILHPFKYKISKKLKSWFEDLEKSDTSVIVGLMVIPINVVNFASDTIGQQFREHYKTVITLLSAIFVLPWLYDSIPRGLSCLFWQLISFISFSTKMLISICTSYITLTIVLAYIISVYTLWTPQRENMFRVTSQLLWLVITSFMASLTGSIQVYTFIFLQIICITGFIMEILDVHKKILLKDPEASIIISIHRVITNSHREKETNTEDSKSDSAQISASETEGTPDKETPSPGTPETLPKSGSWTSNEPSQCPPRPPKILYKTRTLSALPLSGTGKVKQEYRFLPSIKHKSLDDYYMKNNFDNDEETALYFKLLFFGFLCMLVWKNIWLLPLVLVFLVIHFMKIGLDYFGVWLFFENQYNNVALKIKAWWNNRETALIPAHIRGVGKMMSICNTSLRDMAYSSVDTVSSCIVIIGLILFLTVATIFICIQIYSEAIVVVQLSGSLLNSTYVQNSVLHNYLPEGWEEKLDSLIDDAYTYGREGISTTVKNILKEGDPEKVSRVEQQVLEIWDRVYQSWASGHFATAVGPQVDSTAIQDSWDNFVNNVSDSSSLFDYSGVVSWLQANVGTLSAIASSIWAPLASNVSLLAGSLGAFTSLLLGGGSAIINMFINMIVFFTTLFYLLASSNALYKPVEVITQVQPNFGPRLGIALSVAINQVFRASFKMALFYGLWTWLVHNLFGAKVVYLPSGVRINFTSCQNAGWSCFATKDIVYSCANVKSNDFIIHLKDFYTDDIKSLTVQNCKDVKVVLDCPILQKASSLQKFKVKDCDKLQFISLSNNSPVQTPPEVTIENVKEVVSLPRKFFRSPTTANEVKCSGTASLKSISVVDSYIKSINTRAIYNVTGVSSVEFVNVTIGEIQNQGIEAILGNEDTLFSFANNKIESLRYKGVAVQCTSAFFTENTFRDVLSNSLNVTADNLSVVGNKFKQVNGLILKASSIEISKNEFGTLKSNAFTNVKCLRRNTGRRGFTFALNTVENVEPHSLVFDYASCKTAGASVTYENNKIDCRCRDIDFLDAPTELNSLILDLSFNNTCLSAPCLLPVEIVKLLLERGMCDINLDPQIMCLLYGDRHLANNELTTDEDVTEPTSTFYIIRQADPLDGGASSLTAINKDELLRDSHFNITNRTSIRVVFDSSRDFVETLRGTDSSRKKTTDKVNEEYTNRCVGPQCRNNAVQDKQKALDFYKFIYSQLRQPRANDKKKKKRVLVPQLDAVLAAVLGAAPFLGPYLAGLPAALDVWLQGRPLAAVMLPLVQAAPIAFLDAAVYAEIKDGGHPYVTGLAIAGGIFYLGPEGAILGPLLLCCLMVVLNLSSAFLRDTPTDERAALHSRVRYGAYMY
Summary
Uniprot
Pubmed
EMBL
Proteomes
SUPFAM
SSF51126
SSF51126
Gene 3D
ProteinModelPortal
Ontologies
GO
PANTHER
Topology
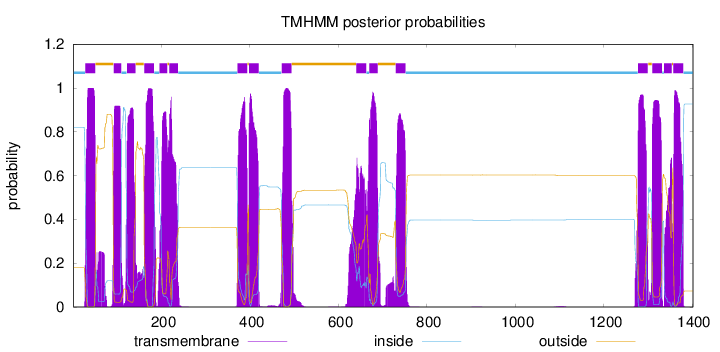
Length:
1402
Number of predicted TMHs:
16
Exp number of AAs in TMHs:
319.591700000001
Exp number, first 60 AAs:
23.96861
Total prob of N-in:
0.82062
POSSIBLE N-term signal
sequence
inside
1 - 27
TMhelix
28 - 50
outside
51 - 91
TMhelix
92 - 109
inside
110 - 121
TMhelix
122 - 141
outside
142 - 160
TMhelix
161 - 183
inside
184 - 195
TMhelix
196 - 213
outside
214 - 217
TMhelix
218 - 237
inside
238 - 371
TMhelix
372 - 394
outside
395 - 397
TMhelix
398 - 420
inside
421 - 471
TMhelix
472 - 494
outside
495 - 640
TMhelix
641 - 663
inside
664 - 669
TMhelix
670 - 689
outside
690 - 729
TMhelix
730 - 752
inside
753 - 1277
TMhelix
1278 - 1300
outside
1301 - 1309
TMhelix
1310 - 1332
inside
1333 - 1336
TMhelix
1337 - 1354
outside
1355 - 1357
TMhelix
1358 - 1380
inside
1381 - 1402
Population Genetic Test Statistics
Pi
229.607716
Theta
193.417375
Tajima's D
0.504901
CLR
0.116191
CSRT
0.519074046297685
Interpretation
Uncertain
