Pre Gene Modal
BGIBMGA004898
Annotation
PREDICTED:_dolichyl-diphosphooligosaccharide--protein_glycosyltransferase_subunit_STT3B_isoform_X1_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.933
Sequence
CDS
ATGCCGTCCACTACTGAGACCCTTCCCAGTAATGGGCGCAGCAAGGGGCTATTTCAAAATACGGCGGGTTTGAGTACGCTCATCACTGTAACTGTCCTGACACTGGCTTGGTTGGCCGGTTTCGCATCCCGGCTTTTCGCTGTGATACGTTTTGAAAGTATAATTCATGAATTCGATCCTTGGTTCAACTACCGGTCTACAGCGTATATGGTTCAGCACGGTTTCTATAATTTTTTGAACTGGTTCGATGAGCGTGCTTGGTACCCACTGGGACGTATCGTGGGGGGTACGGTGTATCCTGGTCTAATGATCACTTCAGGCACTATACATTGGATACTGCATGCTCTCAACATACCCATTCATATCCGAGACATATGTGTATTCCTTGCCCCAGTATTCAGTGGGCTCACAGCAATTGCAACGTACTTATTGACTTCTGAACTATGGTCGCGTGGAGCTGGGCTATTTGCTGCCTGTTTCATAGCTATAGTGCCAGGTTACAGCAGTAGATCCGTTGCTGGGAGCTACGATAATGAAGGAATTGCTATATTTGCATTGCAATTCACATATTACTTGTGGCTGAAAAGTGTTAAAAGTGGGTCCGTATTCTGGTCTATTTGCACTGCATTGTCATATTTTTACATGGTTTCAGCATGGGGCGGATACGTATTCATTATCAATTTGATACCTCTTCACGTCTTTGTTCTTCTAATAATGGGACGTTTCTCGCAGCGTCTTTTCGTCAGCTACTCAGTTTTCTACATTATTGGCTTATTGCTGTCCATGCAGATTCCCTTTGTTGGGTTTCAACCAATACGTACCAGTGAACATATGGCAGCGTCAGGTGTGTTTGCATTGTTGATGGCCATCGGAGCCCTAAAGTACCTGCACAGTCTGAACCCTAAAGGACAATGGAAACAACTGCTAATCTTGGGCGGTATGCTGGCCGCGGGACTCGTGTTCCTGGCTGTGGTGCTCCTCACATACATGGGAGTCATTGCACCTTGGAGTGGAAGGTTTTATTCACTCTGGGACACGGCATACGCAAAGATTCACATTCCGATCATAGCGTCCGTATCCGAGCACCAGCCGACGACGTGGTCTTCATTTTTCTTCGATCTGCACGTGCTCGTGTGCACGTTCCCGGTCGGTCTGTGGTACTGCATCAAGAATGTGAATGATGAGAGAGTTTTCGTTGCTCTGTATGCCCTGAGTGCGGTGTATTTCGCCGGTGTGATGGTTCGTTTGATGTTGACCCTGACGCCCGTAGTGTGCGTGCTAGCCGGCATTGCCTTTTCCATTCTTCTAGACTTGGCCTTAAGAGAGGACGAAGTGCCTGAACAATCCCCGGACAGTGAAGAATCTAAGAGTCTCTATGATAAAGCCGGCAAGGTAAAGAAGCGCACACTTGAGGCCGCGCCGAATGCGGACACCGGCCTGGGCATGAACGTGCGCTCGGGAACGCTCATAGCCTTCATGATCCTGCTGATGTTGTTCTCTGTGCACTGCACCTGGGCCACGTCTAACGCTTACTCCAGCCCTAGCATCGTTCTGGCCAGCTACGGCAATGACGGGTCGCGTAAAATCTTAGATGACTTCCGAGAAGCCTACGGTTGGTTATCGCAGAATACAGCAGAAGACGCAAGGATTATGTCTTGGTGGGACTACGGGTATCAGATAGCGGGGATGGGTAACAGAACAACCCTGGTGGACAATAACACCTGGAACAACTCCCACATAGCACTGGTCGGGAAGGCGATGGCGAGCAACGAATCCGCTGCCTACAGCATCATGACGATGCTGGACGTTGATTACGTGCTCGTTATATTCGGGGGCGCGATCGGGTACTCCGGAGACGACATCAACAAGTTCATATGGATGGTCAGGATCGCTGAGGGGGAACATCCTAAGGACATACACGAAGTTGACTATTTTACTGAAAGAGGAGAATACAGGATCGACTCTGAAGGATCAAAAACTATGTTGAACTGTTTGATGTACAAACTCTCGTATTATAGGTACGACAGTGGGGGAAGCCCTCCCGGGTACGACCGTACGCGCAGTGCGCTGCCCGGGAACAGAGGGTTCAAACTTACCTACCTCGAGGAAGCCTACACCACCGAGCACTGGCTCGTCAGGATATACAGGGTGAAAAAGCCATCTGAGTTCAATCGTCCGCGTTTGTCACTAGCAAAAAGGAAAATTTCAACAAGCAATACTATTTCTAAGAAGACTTCAAGAAGAAGAAAAGGCGCATTGAAGAACAAACCTACAGTCGTGAAGGGAAAAAAGGTCGGTAAACTGGAGTGA
Protein
MPSTTETLPSNGRSKGLFQNTAGLSTLITVTVLTLAWLAGFASRLFAVIRFESIIHEFDPWFNYRSTAYMVQHGFYNFLNWFDERAWYPLGRIVGGTVYPGLMITSGTIHWILHALNIPIHIRDICVFLAPVFSGLTAIATYLLTSELWSRGAGLFAACFIAIVPGYSSRSVAGSYDNEGIAIFALQFTYYLWLKSVKSGSVFWSICTALSYFYMVSAWGGYVFIINLIPLHVFVLLIMGRFSQRLFVSYSVFYIIGLLLSMQIPFVGFQPIRTSEHMAASGVFALLMAIGALKYLHSLNPKGQWKQLLILGGMLAAGLVFLAVVLLTYMGVIAPWSGRFYSLWDTAYAKIHIPIIASVSEHQPTTWSSFFFDLHVLVCTFPVGLWYCIKNVNDERVFVALYALSAVYFAGVMVRLMLTLTPVVCVLAGIAFSILLDLALREDEVPEQSPDSEESKSLYDKAGKVKKRTLEAAPNADTGLGMNVRSGTLIAFMILLMLFSVHCTWATSNAYSSPSIVLASYGNDGSRKILDDFREAYGWLSQNTAEDARIMSWWDYGYQIAGMGNRTTLVDNNTWNNSHIALVGKAMASNESAAYSIMTMLDVDYVLVIFGGAIGYSGDDINKFIWMVRIAEGEHPKDIHEVDYFTERGEYRIDSEGSKTMLNCLMYKLSYYRYDSGGSPPGYDRTRSALPGNRGFKLTYLEEAYTTEHWLVRIYRVKKPSEFNRPRLSLAKRKISTSNTISKKTSRRRKGALKNKPTVVKGKKVGKLE
Summary
Uniprot
H9J5V8
A0A3S2LUY1
A0A194RLG9
A0A194PGR0
A0A212EVS0
D6WTB5
+ More
A0A1B6C951 A0A1B6E412 A0A0S1NJ98 A0A1B6ET97 A0A1B6LG56 A0A1Y1KLK0 K7J1J4 T1I1G9 A0A336M250 A0A0C9R4U4 A0A182JX76 A0A084W1P3 T1PCS9 E2BFZ0 A0A182FL27 A0A1I8MZ91 W8BT95 A0A1Q3FA26 A0A1J1HJ00 A0A026W564 A0A182PA64 A0A182VX60 A0A182GX30 A0A1W4WG99 A0A0N7ZB71 A0A1W4VLR4 A0A0P4WHQ0 B3P6S9 A0A1L8E1Q5 T1DE03 Q17H36 A0A0A9YAQ8 A0A182NA89 A0A2J7R6P5 A0A067QZL7 A0A1I8NNY0 Q9XZ53 B0WNG3 A0A182J1N9 B4PUE5 A0A0K8WJA0 Q7KN63 E2AG81 A0A1Y9HEA9 B3M1Y5 B4IJF1 A0A1Y9J0I6 A0A0L0CCK5 B4JHS0 E0VV86 A0A182QMB9 E9G1S7 A0A182V7H2 A0A182TEE4 Q7QC12 B4LVQ7 U5EI98 A0A182YKH9 A0A1Y9GKN6 A0A182MW20 B4KBG0 B4NJU9 W5J714 A0A340U080 A0A158NCA4 A0A2M3ZE38 A0A2M3ZZW8 A0A2M4BEI2 A0A1A9VM30 A0A1B0CBT8 A0A1A9Y7H2 A0A1B0AKG5 A0A3B0JLV9 B4GE31 B5DU70 A0A1A9W2P2 V9IGH0 F4X834 A0A0M4EWI6 A0A2A3ERQ7 A0A232FJ25 A0A2P2I172 T1J0C4 A0A224XK83 A0A2H8TWJ0 A0A2S2Q9R1 J9JV03 A0A2I9LP13 A0A293LZ14 A0A0L7QM50 A0A2R5L6K1 A0A0J7KTS4 A0A1Z5KVP6 E9JCU0
A0A1B6C951 A0A1B6E412 A0A0S1NJ98 A0A1B6ET97 A0A1B6LG56 A0A1Y1KLK0 K7J1J4 T1I1G9 A0A336M250 A0A0C9R4U4 A0A182JX76 A0A084W1P3 T1PCS9 E2BFZ0 A0A182FL27 A0A1I8MZ91 W8BT95 A0A1Q3FA26 A0A1J1HJ00 A0A026W564 A0A182PA64 A0A182VX60 A0A182GX30 A0A1W4WG99 A0A0N7ZB71 A0A1W4VLR4 A0A0P4WHQ0 B3P6S9 A0A1L8E1Q5 T1DE03 Q17H36 A0A0A9YAQ8 A0A182NA89 A0A2J7R6P5 A0A067QZL7 A0A1I8NNY0 Q9XZ53 B0WNG3 A0A182J1N9 B4PUE5 A0A0K8WJA0 Q7KN63 E2AG81 A0A1Y9HEA9 B3M1Y5 B4IJF1 A0A1Y9J0I6 A0A0L0CCK5 B4JHS0 E0VV86 A0A182QMB9 E9G1S7 A0A182V7H2 A0A182TEE4 Q7QC12 B4LVQ7 U5EI98 A0A182YKH9 A0A1Y9GKN6 A0A182MW20 B4KBG0 B4NJU9 W5J714 A0A340U080 A0A158NCA4 A0A2M3ZE38 A0A2M3ZZW8 A0A2M4BEI2 A0A1A9VM30 A0A1B0CBT8 A0A1A9Y7H2 A0A1B0AKG5 A0A3B0JLV9 B4GE31 B5DU70 A0A1A9W2P2 V9IGH0 F4X834 A0A0M4EWI6 A0A2A3ERQ7 A0A232FJ25 A0A2P2I172 T1J0C4 A0A224XK83 A0A2H8TWJ0 A0A2S2Q9R1 J9JV03 A0A2I9LP13 A0A293LZ14 A0A0L7QM50 A0A2R5L6K1 A0A0J7KTS4 A0A1Z5KVP6 E9JCU0
Pubmed
19121390
26354079
22118469
18362917
19820115
28004739
+ More
20075255 24438588 20798317 25315136 24495485 24508170 30249741 26483478 17994087 24330624 17510324 25401762 24845553 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 26108605 20566863 21292972 12364791 14747013 17210077 25244985 20920257 23761445 21347285 15632085 21719571 28648823 29248469 28528879 21282665
20075255 24438588 20798317 25315136 24495485 24508170 30249741 26483478 17994087 24330624 17510324 25401762 24845553 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 26108605 20566863 21292972 12364791 14747013 17210077 25244985 20920257 23761445 21347285 15632085 21719571 28648823 29248469 28528879 21282665
EMBL
BABH01030048
RSAL01000006
RVE54323.1
KQ460045
KPJ18264.1
KQ459604
+ More
KPI92228.1 AGBW02012127 OWR45595.1 KQ971352 EFA06314.1 GEDC01027272 JAS10026.1 GEDC01004622 JAS32676.1 KT362368 ALL54974.1 GECZ01028593 GECZ01023734 GECZ01021305 GECZ01019067 GECZ01018073 GECZ01011846 GECZ01008984 JAS41176.1 JAS46035.1 JAS48464.1 JAS50702.1 JAS51696.1 JAS57923.1 JAS60785.1 GEBQ01017296 JAT22681.1 GEZM01080230 JAV62283.1 ACPB03016273 UFQT01000309 SSX23079.1 GBYB01002990 JAG72757.1 ATLV01019399 KE525269 KFB44137.1 KA646561 AFP61190.1 GL448096 EFN85392.1 GAMC01004143 JAC02413.1 GFDL01010692 JAV24353.1 CVRI01000006 CRK87989.1 KK107405 QOIP01000014 EZA51220.1 RLU14762.1 JXUM01094529 JXUM01094530 JXUM01094531 JXUM01094532 JXUM01094533 JXUM01094534 JXUM01094535 KQ564135 KXJ72708.1 GDRN01088576 JAI60832.1 GDRN01088577 JAI60831.1 CH954182 EDV53749.1 GFDF01001582 JAV12502.1 GALA01001242 JAA93610.1 CH477253 EAT45977.1 GBHO01014888 GBHO01014886 JAG28716.1 JAG28718.1 NEVH01006756 PNF36504.1 KK853153 KDR10570.1 AE014297 AAF56391.1 DS232010 EDS31617.1 CM000160 EDW98832.1 GDHF01001165 JAI51149.1 AF132552 AAD27851.2 GL439266 EFN67572.1 CH902617 EDV42245.1 CH480848 EDW51142.1 JRES01000595 KNC29985.1 CH916369 EDV93909.1 DS235804 EEB17292.1 AXCN02000218 GL732529 EFX86763.1 AAAB01008859 EAA08102.4 CH940650 EDW66480.1 GANO01002714 JAB57157.1 APCN01000421 AXCM01010392 CH933806 EDW13627.1 CH964272 EDW83951.1 ADMH02002130 ETN58574.1 CCAG010012076 ADTU01011776 GGFM01005990 MBW26741.1 GGFK01000679 MBW34000.1 GGFJ01002319 MBW51460.1 AJWK01005824 JXJN01025262 OUUW01000007 SPP83247.1 CH479182 EDW33866.1 CH672826 EDY71890.1 JR046220 AEY60155.1 GL888912 EGI57383.1 CP012526 ALC47917.1 KZ288194 PBC33809.1 NNAY01000155 OXU30479.1 IACF01002106 LAB67774.1 JH431734 GFTR01007873 JAW08553.1 GFXV01006812 MBW18617.1 GGMS01005275 MBY74478.1 ABLF02016087 GFWZ01000137 MBW20127.1 GFWV01008362 MAA33091.1 KQ414894 KOC59692.1 GGLE01000977 MBY05103.1 LBMM01003343 KMQ93634.1 GFJQ02007922 JAV99047.1 GL771864 EFZ09359.1
KPI92228.1 AGBW02012127 OWR45595.1 KQ971352 EFA06314.1 GEDC01027272 JAS10026.1 GEDC01004622 JAS32676.1 KT362368 ALL54974.1 GECZ01028593 GECZ01023734 GECZ01021305 GECZ01019067 GECZ01018073 GECZ01011846 GECZ01008984 JAS41176.1 JAS46035.1 JAS48464.1 JAS50702.1 JAS51696.1 JAS57923.1 JAS60785.1 GEBQ01017296 JAT22681.1 GEZM01080230 JAV62283.1 ACPB03016273 UFQT01000309 SSX23079.1 GBYB01002990 JAG72757.1 ATLV01019399 KE525269 KFB44137.1 KA646561 AFP61190.1 GL448096 EFN85392.1 GAMC01004143 JAC02413.1 GFDL01010692 JAV24353.1 CVRI01000006 CRK87989.1 KK107405 QOIP01000014 EZA51220.1 RLU14762.1 JXUM01094529 JXUM01094530 JXUM01094531 JXUM01094532 JXUM01094533 JXUM01094534 JXUM01094535 KQ564135 KXJ72708.1 GDRN01088576 JAI60832.1 GDRN01088577 JAI60831.1 CH954182 EDV53749.1 GFDF01001582 JAV12502.1 GALA01001242 JAA93610.1 CH477253 EAT45977.1 GBHO01014888 GBHO01014886 JAG28716.1 JAG28718.1 NEVH01006756 PNF36504.1 KK853153 KDR10570.1 AE014297 AAF56391.1 DS232010 EDS31617.1 CM000160 EDW98832.1 GDHF01001165 JAI51149.1 AF132552 AAD27851.2 GL439266 EFN67572.1 CH902617 EDV42245.1 CH480848 EDW51142.1 JRES01000595 KNC29985.1 CH916369 EDV93909.1 DS235804 EEB17292.1 AXCN02000218 GL732529 EFX86763.1 AAAB01008859 EAA08102.4 CH940650 EDW66480.1 GANO01002714 JAB57157.1 APCN01000421 AXCM01010392 CH933806 EDW13627.1 CH964272 EDW83951.1 ADMH02002130 ETN58574.1 CCAG010012076 ADTU01011776 GGFM01005990 MBW26741.1 GGFK01000679 MBW34000.1 GGFJ01002319 MBW51460.1 AJWK01005824 JXJN01025262 OUUW01000007 SPP83247.1 CH479182 EDW33866.1 CH672826 EDY71890.1 JR046220 AEY60155.1 GL888912 EGI57383.1 CP012526 ALC47917.1 KZ288194 PBC33809.1 NNAY01000155 OXU30479.1 IACF01002106 LAB67774.1 JH431734 GFTR01007873 JAW08553.1 GFXV01006812 MBW18617.1 GGMS01005275 MBY74478.1 ABLF02016087 GFWZ01000137 MBW20127.1 GFWV01008362 MAA33091.1 KQ414894 KOC59692.1 GGLE01000977 MBY05103.1 LBMM01003343 KMQ93634.1 GFJQ02007922 JAV99047.1 GL771864 EFZ09359.1
Proteomes
UP000005204
UP000283053
UP000053240
UP000053268
UP000007151
UP000007266
+ More
UP000002358 UP000015103 UP000075881 UP000030765 UP000008237 UP000069272 UP000095301 UP000183832 UP000053097 UP000279307 UP000075885 UP000075920 UP000069940 UP000249989 UP000192223 UP000192221 UP000008711 UP000008820 UP000075884 UP000235965 UP000027135 UP000095300 UP000000803 UP000002320 UP000075880 UP000002282 UP000000311 UP000075900 UP000007801 UP000001292 UP000076407 UP000037069 UP000001070 UP000009046 UP000075886 UP000000305 UP000075903 UP000075902 UP000007062 UP000008792 UP000076408 UP000075840 UP000075883 UP000009192 UP000007798 UP000000673 UP000092444 UP000005205 UP000078200 UP000092461 UP000092443 UP000092460 UP000268350 UP000008744 UP000001819 UP000091820 UP000007755 UP000092553 UP000242457 UP000215335 UP000007819 UP000053825 UP000036403
UP000002358 UP000015103 UP000075881 UP000030765 UP000008237 UP000069272 UP000095301 UP000183832 UP000053097 UP000279307 UP000075885 UP000075920 UP000069940 UP000249989 UP000192223 UP000192221 UP000008711 UP000008820 UP000075884 UP000235965 UP000027135 UP000095300 UP000000803 UP000002320 UP000075880 UP000002282 UP000000311 UP000075900 UP000007801 UP000001292 UP000076407 UP000037069 UP000001070 UP000009046 UP000075886 UP000000305 UP000075903 UP000075902 UP000007062 UP000008792 UP000076408 UP000075840 UP000075883 UP000009192 UP000007798 UP000000673 UP000092444 UP000005205 UP000078200 UP000092461 UP000092443 UP000092460 UP000268350 UP000008744 UP000001819 UP000091820 UP000007755 UP000092553 UP000242457 UP000215335 UP000007819 UP000053825 UP000036403
Pfam
Interpro
IPR003674
Oligo_trans_STT3
+ More
IPR002110 Ankyrin_rpt
IPR027475 Asparaginase/glutaminase_AS2
IPR006033 AsnA_fam
IPR041725 L-asparaginase_I
IPR040919 Asparaginase_C
IPR027473 L-asparaginase_C
IPR027474 L-asparaginase_N
IPR037152 L-asparaginase_N_sf
IPR036152 Asp/glu_Ase-like_sf
IPR020683 Ankyrin_rpt-contain_dom
IPR006034 Asparaginase/glutaminase-like
IPR036770 Ankyrin_rpt-contain_sf
IPR031949 DUF4776
IPR000086 NUDIX_hydrolase_dom
IPR011876 IsopentenylPP_isomerase_typ1
IPR015797 NUDIX_hydrolase-like_dom_sf
IPR002110 Ankyrin_rpt
IPR027475 Asparaginase/glutaminase_AS2
IPR006033 AsnA_fam
IPR041725 L-asparaginase_I
IPR040919 Asparaginase_C
IPR027473 L-asparaginase_C
IPR027474 L-asparaginase_N
IPR037152 L-asparaginase_N_sf
IPR036152 Asp/glu_Ase-like_sf
IPR020683 Ankyrin_rpt-contain_dom
IPR006034 Asparaginase/glutaminase-like
IPR036770 Ankyrin_rpt-contain_sf
IPR031949 DUF4776
IPR000086 NUDIX_hydrolase_dom
IPR011876 IsopentenylPP_isomerase_typ1
IPR015797 NUDIX_hydrolase-like_dom_sf
Gene 3D
ProteinModelPortal
H9J5V8
A0A3S2LUY1
A0A194RLG9
A0A194PGR0
A0A212EVS0
D6WTB5
+ More
A0A1B6C951 A0A1B6E412 A0A0S1NJ98 A0A1B6ET97 A0A1B6LG56 A0A1Y1KLK0 K7J1J4 T1I1G9 A0A336M250 A0A0C9R4U4 A0A182JX76 A0A084W1P3 T1PCS9 E2BFZ0 A0A182FL27 A0A1I8MZ91 W8BT95 A0A1Q3FA26 A0A1J1HJ00 A0A026W564 A0A182PA64 A0A182VX60 A0A182GX30 A0A1W4WG99 A0A0N7ZB71 A0A1W4VLR4 A0A0P4WHQ0 B3P6S9 A0A1L8E1Q5 T1DE03 Q17H36 A0A0A9YAQ8 A0A182NA89 A0A2J7R6P5 A0A067QZL7 A0A1I8NNY0 Q9XZ53 B0WNG3 A0A182J1N9 B4PUE5 A0A0K8WJA0 Q7KN63 E2AG81 A0A1Y9HEA9 B3M1Y5 B4IJF1 A0A1Y9J0I6 A0A0L0CCK5 B4JHS0 E0VV86 A0A182QMB9 E9G1S7 A0A182V7H2 A0A182TEE4 Q7QC12 B4LVQ7 U5EI98 A0A182YKH9 A0A1Y9GKN6 A0A182MW20 B4KBG0 B4NJU9 W5J714 A0A340U080 A0A158NCA4 A0A2M3ZE38 A0A2M3ZZW8 A0A2M4BEI2 A0A1A9VM30 A0A1B0CBT8 A0A1A9Y7H2 A0A1B0AKG5 A0A3B0JLV9 B4GE31 B5DU70 A0A1A9W2P2 V9IGH0 F4X834 A0A0M4EWI6 A0A2A3ERQ7 A0A232FJ25 A0A2P2I172 T1J0C4 A0A224XK83 A0A2H8TWJ0 A0A2S2Q9R1 J9JV03 A0A2I9LP13 A0A293LZ14 A0A0L7QM50 A0A2R5L6K1 A0A0J7KTS4 A0A1Z5KVP6 E9JCU0
A0A1B6C951 A0A1B6E412 A0A0S1NJ98 A0A1B6ET97 A0A1B6LG56 A0A1Y1KLK0 K7J1J4 T1I1G9 A0A336M250 A0A0C9R4U4 A0A182JX76 A0A084W1P3 T1PCS9 E2BFZ0 A0A182FL27 A0A1I8MZ91 W8BT95 A0A1Q3FA26 A0A1J1HJ00 A0A026W564 A0A182PA64 A0A182VX60 A0A182GX30 A0A1W4WG99 A0A0N7ZB71 A0A1W4VLR4 A0A0P4WHQ0 B3P6S9 A0A1L8E1Q5 T1DE03 Q17H36 A0A0A9YAQ8 A0A182NA89 A0A2J7R6P5 A0A067QZL7 A0A1I8NNY0 Q9XZ53 B0WNG3 A0A182J1N9 B4PUE5 A0A0K8WJA0 Q7KN63 E2AG81 A0A1Y9HEA9 B3M1Y5 B4IJF1 A0A1Y9J0I6 A0A0L0CCK5 B4JHS0 E0VV86 A0A182QMB9 E9G1S7 A0A182V7H2 A0A182TEE4 Q7QC12 B4LVQ7 U5EI98 A0A182YKH9 A0A1Y9GKN6 A0A182MW20 B4KBG0 B4NJU9 W5J714 A0A340U080 A0A158NCA4 A0A2M3ZE38 A0A2M3ZZW8 A0A2M4BEI2 A0A1A9VM30 A0A1B0CBT8 A0A1A9Y7H2 A0A1B0AKG5 A0A3B0JLV9 B4GE31 B5DU70 A0A1A9W2P2 V9IGH0 F4X834 A0A0M4EWI6 A0A2A3ERQ7 A0A232FJ25 A0A2P2I172 T1J0C4 A0A224XK83 A0A2H8TWJ0 A0A2S2Q9R1 J9JV03 A0A2I9LP13 A0A293LZ14 A0A0L7QM50 A0A2R5L6K1 A0A0J7KTS4 A0A1Z5KVP6 E9JCU0
PDB
6FTI
E-value=0,
Score=2011
Ontologies
PATHWAY
GO
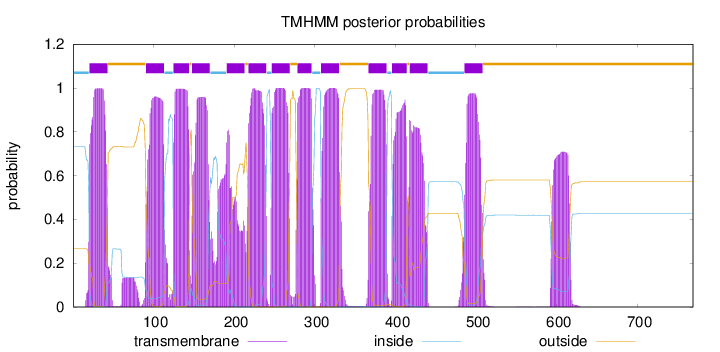
Topology
Length:
769
Number of predicted TMHs:
13
Exp number of AAs in TMHs:
285.696109999999
Exp number, first 60 AAs:
22.14802
Total prob of N-in:
0.73231
POSSIBLE N-term signal
sequence
inside
1 - 20
TMhelix
21 - 43
outside
44 - 90
TMhelix
91 - 113
inside
114 - 124
TMhelix
125 - 144
outside
145 - 147
TMhelix
148 - 170
inside
171 - 190
TMhelix
191 - 213
outside
214 - 217
TMhelix
218 - 240
inside
241 - 246
TMhelix
247 - 269
outside
270 - 278
TMhelix
279 - 296
inside
297 - 307
TMhelix
308 - 330
outside
331 - 366
TMhelix
367 - 389
inside
390 - 395
TMhelix
396 - 414
outside
415 - 417
TMhelix
418 - 440
inside
441 - 485
TMhelix
486 - 508
outside
509 - 769
Population Genetic Test Statistics
Pi
288.752573
Theta
205.669301
Tajima's D
0.665528
CLR
0.66621
CSRT
0.565521723913804
Interpretation
Uncertain
