Gene
KWMTBOMO15098
Pre Gene Modal
BGIBMGA005003
Annotation
hypothetical_protein_KGM_19935_[Danaus_plexippus]
Location in the cell
Extracellular Reliability : 2.951
Sequence
CDS
ATGCCTAGTGCCTCGTTTTCGTCTTCGCGTTCTTACGTACGTAGATCGCGGAGAAAAGGCGGTCATCGAATACCACTACCTTCTAGGACACAATCGAGTGAACCATGTGAGTCAGTTCCAATACCCAACAACGTCACGGGCAGTGCGATGGCGGCGACGGCCGCGTGTGCTGGCCCAAGCGGTGGGGGCGGAGCACCACCCGTACCTGCTGCTGCACCCGCCACCACCGCCGGCCATCATAGCCCTTACGACCTGCGCCGTAAGTCGCCACCAGCTTATCACGAGCCCGGACCTTCAGGTGCTTGCTCATTGCCGGCCAGGAAGAGACCTAGAACATCACTCTCTCAAGGCGTGGACGTGTGCAATGTCTCGCAGTACTTGCAGTATGAGCTGCCGGATGAAGTGTTGCTCTGTATCCTGTCATACTTGACTGAACGCGACCTGTGCCGTGTCGCTCAAGTCTGCAAGAGATTTAATACAATCGCGAATGATACTGAACTATGGAAAAGTTTATATCAGTCTGTGTTTGAGTATGACACTCCACTGATGCACCCAGCACCGTGCCAGTTTGAGTTTGTAGCGCCTGATGAGTGCGACGCAGACAACCCATGGAAGGAGAGCTTTCGTCAGCTCTACTATGGCATACACGTCCGCCCTAGTTTTCGTCTAAAGAAGGATTCGCGTATCAGGCATTTTAATACAATCAGGACTGCACTGGAATTCGTGGAGGAGCGGACAGGCAGCAGTGTAACCGCTAGTACGAATACCGCGGGCCCCACCACGAGCAGCTCTAGCAGTAGTGCCGCAAGCACAACCAACACCAGCAACAGTAACGTGTGCTCGTGCAACCCCACTGGCGCTTGTACGTGTCGCCGCCCGCAACCAACCCACCAACCGGCGCTAGTCTTCATACACGCTGGACTCTATCAAGAGGAATGTCTTGCTATTGATTCTGATGTGCAGCTTATTGGTTGTGCGACGGGCAATGTAGCGGAGTCTGTAGTACTTGAGCGTGAAGCGGAATCGACGCTGACGTTTGCGGAGGGAGCGACTCGGGCTTATGCCGGACACATGACGCTCAAGTTCTCACCGGATGCAACAAGTACCATGCAGCATCATAAGCACTTCTGTCTCGAGGTCTCAGATAACTGTTCACCCACAATTGATCACTGTATTATCAGGAGCGCTAGTGTTGTGGGTGCAGCAGTGTGCGTTAGCGGTGCCGGCGCGAATCCCGTAATCAAGCACTGTGACATTAGTGACTGCGAGAACGTTGGTCTCTATGTCACAGACTACGCGCAAGGCGCTTATCAGGATAATGAAATCTCAAGAAACGCCCTTGCTGGCATATGGGTCAAAAATTTTGCCAACCCTATTATGCGTCGTAACCATATACATCACGGACGTGACGTCGGTATATTTACGTTCGAGAATGGATTGGGTTACTTTGAAGCCAACGATATTCACAACAATAGAATCGCCGGTTTCGAAGTTAAGGCCGGCGCGAACCCCACCGTAGTGCACTGCGAGATCCACCACGGTCAAACTGGTGGTATTTATGTGCACGAATCTGGTTTAGGCCAATTTATTGATAACAAGATACATTCGAATAACTTTGCCGGCGTATGGATAACGTCCAACAGCAACCCCACCATACGGCGTAATGAAATATATAATGGGCATCAAGGAGGCGTGTACATATTTGGCGAAGGACGAGGTCTCATTGAACATAATAATATATATGGAAATGCTCTTGCTGGTATACAGATACGCACAAACAGTGATCCTATTGTGAGACATAATAAGATTCATCACGGGCAGCATGGAGGTATATATGTACATGAAAAAGGTCAAGGACTCATTGAAGAGAATGAAGTTTATGCCAACACATTAGCTGGTGTTTGGATAACTACTGGTTCCACACCTGTTCTGCGCCGCAACCGTATACATTCAGGAAAACAGGTTGGTGTATACTTCTATGACAATGGTCACGGCAAGTTAGAAGACAATGACATATTCAACCACCTATACTCTGGAGTTCAGATTAGGACAGGCAGCAACCCTGTTATCAGAGGAAACAAAATTTGGGGTGGTCAGAATGGTGGTGTACTTGTATACAACGGGGGCCTCGGATTGTTGGAGCAGAACGAGATATTCGATAATGCGATGGCCGGTGTATGGATTAAGACTGATTCAAACCCAACGCTCAAGAGGAATAAGATCTTCGACGGACGTGACGGGGGCATTTGTATTTTTAATGGTGGGAAGGGAGTGTTAGAAGAGAATGACATATTTCACAACGCGCAAGCAGGCGTCCTGATCTCAACGCAGAGCCATCCAGTATTACGTCGGAACAGAATCTTTGACGGACTGGCCGCCGGTGTCGAGATTACTAATAATGCAACCGCCACCCTAGAACATAATCAGATATTTAATAACAGATTCGGAGGCCTGTGTCTGGCTTCTGGCGTTACTCCTTTAGTTAGGGGAAATAAGATATTCAGCAACCAAGACGCCGTTGAAAAGGCTGTCGGCGGTGGACAGTGTCTCTACAAAATCTCTTCTTATACTTCCTTTCCGATGCACGATTTCTATAGGTGCCAGACGTGCAATACGACAGACCGCAATGCAATCTGCGTGAACTGCATCAAAACGTGCCATTCCGGTCATGACGTCGAATTTATTCGTCATGACAGGTTCTTCTGCGACTGCGGCGCGGGCACGCTCTCGAACCAGTGCCAGCTGCAGGGCGAGCCCACGCAGGACACGGACACGCTGTACGACTCGGCCGCACCCATGGAGTCGCACACGCTGATGGTCAACTGA
Protein
MPSASFSSSRSYVRRSRRKGGHRIPLPSRTQSSEPCESVPIPNNVTGSAMAATAACAGPSGGGGAPPVPAAAPATTAGHHSPYDLRRKSPPAYHEPGPSGACSLPARKRPRTSLSQGVDVCNVSQYLQYELPDEVLLCILSYLTERDLCRVAQVCKRFNTIANDTELWKSLYQSVFEYDTPLMHPAPCQFEFVAPDECDADNPWKESFRQLYYGIHVRPSFRLKKDSRIRHFNTIRTALEFVEERTGSSVTASTNTAGPTTSSSSSSAASTTNTSNSNVCSCNPTGACTCRRPQPTHQPALVFIHAGLYQEECLAIDSDVQLIGCATGNVAESVVLEREAESTLTFAEGATRAYAGHMTLKFSPDATSTMQHHKHFCLEVSDNCSPTIDHCIIRSASVVGAAVCVSGAGANPVIKHCDISDCENVGLYVTDYAQGAYQDNEISRNALAGIWVKNFANPIMRRNHIHHGRDVGIFTFENGLGYFEANDIHNNRIAGFEVKAGANPTVVHCEIHHGQTGGIYVHESGLGQFIDNKIHSNNFAGVWITSNSNPTIRRNEIYNGHQGGVYIFGEGRGLIEHNNIYGNALAGIQIRTNSDPIVRHNKIHHGQHGGIYVHEKGQGLIEENEVYANTLAGVWITTGSTPVLRRNRIHSGKQVGVYFYDNGHGKLEDNDIFNHLYSGVQIRTGSNPVIRGNKIWGGQNGGVLVYNGGLGLLEQNEIFDNAMAGVWIKTDSNPTLKRNKIFDGRDGGICIFNGGKGVLEENDIFHNAQAGVLISTQSHPVLRRNRIFDGLAAGVEITNNATATLEHNQIFNNRFGGLCLASGVTPLVRGNKIFSNQDAVEKAVGGGQCLYKISSYTSFPMHDFYRCQTCNTTDRNAICVNCIKTCHSGHDVEFIRHDRFFCDCGAGTLSNQCQLQGEPTQDTDTLYDSAAPMESHTLMVN
Summary
Uniprot
A0A2H1VTA8
A0A212ER61
A0A194PM84
H9J662
A0A194RL46
A0A2A4JD23
+ More
V5GXU9 A0A1Y1M2Y7 A0A1W4WWX3 A0A1W4X6A8 A0A2A3EQB8 A0A087ZZQ0 A0A0M8ZSU5 E9IBF2 A0A0L7R1D2 A0A026WIB9 F4WG38 A0A158NWK6 E2AKZ9 E2BN51 A0A232FGJ7 D6WSA1 A0A195C6M8 A0A151XBS1 A0A195EZM6 A0A195EFG6 A0A195BHA1 A0A154PHF5 U4UQT2 E0VNH3 A0A1B6E3W9 A0A0C9PSS7 A0A1B0CDH5 A0A1L8DAX7 A0A067QJS4 A0A1B6J9Y3 A0A1B6ELU9 A0A0A9Y110 B4HJD0 A0A2P8Z2Q5 A0A1I8PNX3 B3P4P6 B4QVX6 W8BGN1 Q9VH60 A0A0K8SQ96 A0A0A1WPR0 Q6NQY0 B4PVG4 A0A1A9X5Y2 A0A1A9V5I5 A0A1B0ASN7 A0A1Q3FVQ7 A0A1Q3FVJ8 A0A1Q3FVJ0 A0A1Q3FVK2 A0A1A9Z745 B4JF41 B4M170 A0A0K8UV86 A0A0P6BQ65 E9HAQ2 A0A0M4ENN6 A0A1W4W070 A0A1I8N9L3 Q299I8 B4G581 A0A0P5PGB4 A0A164YID3 A0A1S4F8W1 A0A0P6ISR2 A0A336LV74 B3M1D3 A0A3B0KCU5 U5EWF2 A0A0P4VN79 B4NKK3 A0A1B0G2I3 A0A0L0CGP8 A0A224X717 A0A069DZL3 A0A023F5L5 B4K952 A0A1J1I2Y4 A0A0P5E1B8 A0A131XXE4 A0A147BWB8 T1HZG2 L7MF40 A0A1E1XB69 A0A224YMR3 A0A3S3NW13 A0A3S4QNS7 T1IYN2 V5H2M0 A0A2S2R020 A0A023GLY2 A0A1E1XQG1 L7MKH5 J9K0E1
V5GXU9 A0A1Y1M2Y7 A0A1W4WWX3 A0A1W4X6A8 A0A2A3EQB8 A0A087ZZQ0 A0A0M8ZSU5 E9IBF2 A0A0L7R1D2 A0A026WIB9 F4WG38 A0A158NWK6 E2AKZ9 E2BN51 A0A232FGJ7 D6WSA1 A0A195C6M8 A0A151XBS1 A0A195EZM6 A0A195EFG6 A0A195BHA1 A0A154PHF5 U4UQT2 E0VNH3 A0A1B6E3W9 A0A0C9PSS7 A0A1B0CDH5 A0A1L8DAX7 A0A067QJS4 A0A1B6J9Y3 A0A1B6ELU9 A0A0A9Y110 B4HJD0 A0A2P8Z2Q5 A0A1I8PNX3 B3P4P6 B4QVX6 W8BGN1 Q9VH60 A0A0K8SQ96 A0A0A1WPR0 Q6NQY0 B4PVG4 A0A1A9X5Y2 A0A1A9V5I5 A0A1B0ASN7 A0A1Q3FVQ7 A0A1Q3FVJ8 A0A1Q3FVJ0 A0A1Q3FVK2 A0A1A9Z745 B4JF41 B4M170 A0A0K8UV86 A0A0P6BQ65 E9HAQ2 A0A0M4ENN6 A0A1W4W070 A0A1I8N9L3 Q299I8 B4G581 A0A0P5PGB4 A0A164YID3 A0A1S4F8W1 A0A0P6ISR2 A0A336LV74 B3M1D3 A0A3B0KCU5 U5EWF2 A0A0P4VN79 B4NKK3 A0A1B0G2I3 A0A0L0CGP8 A0A224X717 A0A069DZL3 A0A023F5L5 B4K952 A0A1J1I2Y4 A0A0P5E1B8 A0A131XXE4 A0A147BWB8 T1HZG2 L7MF40 A0A1E1XB69 A0A224YMR3 A0A3S3NW13 A0A3S4QNS7 T1IYN2 V5H2M0 A0A2S2R020 A0A023GLY2 A0A1E1XQG1 L7MKH5 J9K0E1
Pubmed
22118469
26354079
19121390
28004739
21282665
24508170
+ More
30249741 21719571 21347285 20798317 28648823 18362917 19820115 23537049 20566863 24845553 25401762 26823975 17994087 29403074 24495485 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25830018 17550304 21292972 25315136 15632085 26999592 27129103 26108605 26334808 25474469 29652888 25576852 28503490 28797301 25765539 29209593
30249741 21719571 21347285 20798317 28648823 18362917 19820115 23537049 20566863 24845553 25401762 26823975 17994087 29403074 24495485 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25830018 17550304 21292972 25315136 15632085 26999592 27129103 26108605 26334808 25474469 29652888 25576852 28503490 28797301 25765539 29209593
EMBL
ODYU01004018
SOQ43464.1
AGBW02013098
OWR43982.1
KQ459604
KPI92225.1
+ More
BABH01030048 KQ460045 KPJ18267.1 NWSH01002042 PCG69322.1 GALX01003273 JAB65193.1 GEZM01042460 JAV79841.1 KZ288194 PBC33880.1 KQ435863 KOX70425.1 GL762111 EFZ22028.1 KQ414667 KOC64680.1 KK107185 QOIP01000012 EZA55760.1 RLU15695.1 GL888128 EGI66805.1 ADTU01028135 GL440425 EFN65916.1 GL449385 EFN82813.1 NNAY01000255 OXU29683.1 KQ971354 EFA07112.2 KQ978205 KYM96492.1 KQ982320 KYQ57805.1 KQ981897 KYN33673.1 KQ978957 KYN26988.1 KQ976488 KYM83603.1 KQ434905 KZC11237.1 KB632326 ERL92410.1 DS235339 EEB14929.1 GEDC01004665 JAS32633.1 GBYB01004393 JAG74160.1 AJWK01007835 AJWK01007836 GFDF01010502 JAV03582.1 KK853256 KDR09267.1 GECU01011689 JAS96017.1 GECZ01030862 JAS38907.1 GBHO01020430 GBHO01020429 GDHC01014286 JAG23174.1 JAG23175.1 JAQ04343.1 CH480815 EDW42802.1 PYGN01000223 PSN50779.1 CH954181 EDV49699.1 CM000364 EDX13556.1 GAMC01010442 JAB96113.1 AE014297 AAF54459.1 AGB95823.1 GBRD01010346 JAG55478.1 GBXI01013500 JAD00792.1 BT010305 AAQ23623.1 CM000160 EDW96737.1 JXJN01002887 JXJN01002888 GFDL01003417 JAV31628.1 GFDL01003414 JAV31631.1 GFDL01003415 JAV31630.1 GFDL01003416 JAV31629.1 CH916369 EDV93322.1 CH940650 EDW67481.1 GDHF01027306 GDHF01025266 GDHF01021813 GDHF01018596 GDHF01018128 GDHF01015803 GDHF01000512 JAI25008.1 JAI27048.1 JAI30501.1 JAI33718.1 JAI34186.1 JAI36511.1 JAI51802.1 GDIP01011420 JAM92295.1 GL732612 EFX71222.1 CP012526 ALC46338.1 CM000070 EAL27715.2 CH479179 EDW24747.1 GDIQ01128961 JAL22765.1 LRGB01000915 KZS15283.1 GDUN01001133 JAN94786.1 UFQT01000217 SSX21844.1 CH902617 EDV42160.1 OUUW01000007 SPP82831.1 GANO01000605 JAB59266.1 GDKW01000524 JAI56071.1 CH964272 EDW85175.1 CCAG010018315 JRES01000409 KNC31598.1 GFTR01008181 JAW08245.1 GBGD01000440 JAC88449.1 GBBI01002260 JAC16452.1 CH933806 EDW14465.1 CVRI01000039 CRK94645.1 GDIP01148680 JAJ74722.1 GEFM01004449 JAP71347.1 GEGO01000592 JAR94812.1 ACPB03006020 GACK01003230 JAA61804.1 GFAC01002714 JAT96474.1 GFPF01007740 MAA18886.1 NCKU01007020 RWS02959.1 NCKU01004506 RWS05855.1 JH431693 GANP01007233 JAB77235.1 GGMS01014158 MBY83361.1 GBBM01000506 JAC34912.1 GFAA01001882 JAU01553.1 GACK01001375 JAA63659.1 ABLF02018337 ABLF02018338 ABLF02018340 ABLF02018341 ABLF02018342
BABH01030048 KQ460045 KPJ18267.1 NWSH01002042 PCG69322.1 GALX01003273 JAB65193.1 GEZM01042460 JAV79841.1 KZ288194 PBC33880.1 KQ435863 KOX70425.1 GL762111 EFZ22028.1 KQ414667 KOC64680.1 KK107185 QOIP01000012 EZA55760.1 RLU15695.1 GL888128 EGI66805.1 ADTU01028135 GL440425 EFN65916.1 GL449385 EFN82813.1 NNAY01000255 OXU29683.1 KQ971354 EFA07112.2 KQ978205 KYM96492.1 KQ982320 KYQ57805.1 KQ981897 KYN33673.1 KQ978957 KYN26988.1 KQ976488 KYM83603.1 KQ434905 KZC11237.1 KB632326 ERL92410.1 DS235339 EEB14929.1 GEDC01004665 JAS32633.1 GBYB01004393 JAG74160.1 AJWK01007835 AJWK01007836 GFDF01010502 JAV03582.1 KK853256 KDR09267.1 GECU01011689 JAS96017.1 GECZ01030862 JAS38907.1 GBHO01020430 GBHO01020429 GDHC01014286 JAG23174.1 JAG23175.1 JAQ04343.1 CH480815 EDW42802.1 PYGN01000223 PSN50779.1 CH954181 EDV49699.1 CM000364 EDX13556.1 GAMC01010442 JAB96113.1 AE014297 AAF54459.1 AGB95823.1 GBRD01010346 JAG55478.1 GBXI01013500 JAD00792.1 BT010305 AAQ23623.1 CM000160 EDW96737.1 JXJN01002887 JXJN01002888 GFDL01003417 JAV31628.1 GFDL01003414 JAV31631.1 GFDL01003415 JAV31630.1 GFDL01003416 JAV31629.1 CH916369 EDV93322.1 CH940650 EDW67481.1 GDHF01027306 GDHF01025266 GDHF01021813 GDHF01018596 GDHF01018128 GDHF01015803 GDHF01000512 JAI25008.1 JAI27048.1 JAI30501.1 JAI33718.1 JAI34186.1 JAI36511.1 JAI51802.1 GDIP01011420 JAM92295.1 GL732612 EFX71222.1 CP012526 ALC46338.1 CM000070 EAL27715.2 CH479179 EDW24747.1 GDIQ01128961 JAL22765.1 LRGB01000915 KZS15283.1 GDUN01001133 JAN94786.1 UFQT01000217 SSX21844.1 CH902617 EDV42160.1 OUUW01000007 SPP82831.1 GANO01000605 JAB59266.1 GDKW01000524 JAI56071.1 CH964272 EDW85175.1 CCAG010018315 JRES01000409 KNC31598.1 GFTR01008181 JAW08245.1 GBGD01000440 JAC88449.1 GBBI01002260 JAC16452.1 CH933806 EDW14465.1 CVRI01000039 CRK94645.1 GDIP01148680 JAJ74722.1 GEFM01004449 JAP71347.1 GEGO01000592 JAR94812.1 ACPB03006020 GACK01003230 JAA61804.1 GFAC01002714 JAT96474.1 GFPF01007740 MAA18886.1 NCKU01007020 RWS02959.1 NCKU01004506 RWS05855.1 JH431693 GANP01007233 JAB77235.1 GGMS01014158 MBY83361.1 GBBM01000506 JAC34912.1 GFAA01001882 JAU01553.1 GACK01001375 JAA63659.1 ABLF02018337 ABLF02018338 ABLF02018340 ABLF02018341 ABLF02018342
Proteomes
UP000007151
UP000053268
UP000005204
UP000053240
UP000218220
UP000192223
+ More
UP000242457 UP000005203 UP000053105 UP000053825 UP000053097 UP000279307 UP000007755 UP000005205 UP000000311 UP000008237 UP000215335 UP000007266 UP000078542 UP000075809 UP000078541 UP000078492 UP000078540 UP000076502 UP000030742 UP000009046 UP000092461 UP000027135 UP000001292 UP000245037 UP000095300 UP000008711 UP000000304 UP000000803 UP000002282 UP000092443 UP000078200 UP000092460 UP000092445 UP000001070 UP000008792 UP000000305 UP000092553 UP000192221 UP000095301 UP000001819 UP000008744 UP000076858 UP000007801 UP000268350 UP000007798 UP000092444 UP000037069 UP000009192 UP000183832 UP000015103 UP000285301 UP000007819
UP000242457 UP000005203 UP000053105 UP000053825 UP000053097 UP000279307 UP000007755 UP000005205 UP000000311 UP000008237 UP000215335 UP000007266 UP000078542 UP000075809 UP000078541 UP000078492 UP000078540 UP000076502 UP000030742 UP000009046 UP000092461 UP000027135 UP000001292 UP000245037 UP000095300 UP000008711 UP000000304 UP000000803 UP000002282 UP000092443 UP000078200 UP000092460 UP000092445 UP000001070 UP000008792 UP000000305 UP000092553 UP000192221 UP000095301 UP000001819 UP000008744 UP000076858 UP000007801 UP000268350 UP000007798 UP000092444 UP000037069 UP000009192 UP000183832 UP000015103 UP000285301 UP000007819
Interpro
IPR011050
Pectin_lyase_fold/virulence
+ More
IPR006633 Carb-bd_sugar_hydrolysis-dom
IPR006626 PbH1
IPR007742 NosD_dom
IPR012334 Pectin_lyas_fold
IPR022441 Para_beta_helix_rpt-2
IPR036047 F-box-like_dom_sf
IPR001810 F-box_dom
IPR003126 Znf_UBR
IPR039448 Beta_helix
IPR029799 FBX11/DRE-1
IPR023828 Peptidase_S8_Ser-AS
IPR006633 Carb-bd_sugar_hydrolysis-dom
IPR006626 PbH1
IPR007742 NosD_dom
IPR012334 Pectin_lyas_fold
IPR022441 Para_beta_helix_rpt-2
IPR036047 F-box-like_dom_sf
IPR001810 F-box_dom
IPR003126 Znf_UBR
IPR039448 Beta_helix
IPR029799 FBX11/DRE-1
IPR023828 Peptidase_S8_Ser-AS
Gene 3D
ProteinModelPortal
A0A2H1VTA8
A0A212ER61
A0A194PM84
H9J662
A0A194RL46
A0A2A4JD23
+ More
V5GXU9 A0A1Y1M2Y7 A0A1W4WWX3 A0A1W4X6A8 A0A2A3EQB8 A0A087ZZQ0 A0A0M8ZSU5 E9IBF2 A0A0L7R1D2 A0A026WIB9 F4WG38 A0A158NWK6 E2AKZ9 E2BN51 A0A232FGJ7 D6WSA1 A0A195C6M8 A0A151XBS1 A0A195EZM6 A0A195EFG6 A0A195BHA1 A0A154PHF5 U4UQT2 E0VNH3 A0A1B6E3W9 A0A0C9PSS7 A0A1B0CDH5 A0A1L8DAX7 A0A067QJS4 A0A1B6J9Y3 A0A1B6ELU9 A0A0A9Y110 B4HJD0 A0A2P8Z2Q5 A0A1I8PNX3 B3P4P6 B4QVX6 W8BGN1 Q9VH60 A0A0K8SQ96 A0A0A1WPR0 Q6NQY0 B4PVG4 A0A1A9X5Y2 A0A1A9V5I5 A0A1B0ASN7 A0A1Q3FVQ7 A0A1Q3FVJ8 A0A1Q3FVJ0 A0A1Q3FVK2 A0A1A9Z745 B4JF41 B4M170 A0A0K8UV86 A0A0P6BQ65 E9HAQ2 A0A0M4ENN6 A0A1W4W070 A0A1I8N9L3 Q299I8 B4G581 A0A0P5PGB4 A0A164YID3 A0A1S4F8W1 A0A0P6ISR2 A0A336LV74 B3M1D3 A0A3B0KCU5 U5EWF2 A0A0P4VN79 B4NKK3 A0A1B0G2I3 A0A0L0CGP8 A0A224X717 A0A069DZL3 A0A023F5L5 B4K952 A0A1J1I2Y4 A0A0P5E1B8 A0A131XXE4 A0A147BWB8 T1HZG2 L7MF40 A0A1E1XB69 A0A224YMR3 A0A3S3NW13 A0A3S4QNS7 T1IYN2 V5H2M0 A0A2S2R020 A0A023GLY2 A0A1E1XQG1 L7MKH5 J9K0E1
V5GXU9 A0A1Y1M2Y7 A0A1W4WWX3 A0A1W4X6A8 A0A2A3EQB8 A0A087ZZQ0 A0A0M8ZSU5 E9IBF2 A0A0L7R1D2 A0A026WIB9 F4WG38 A0A158NWK6 E2AKZ9 E2BN51 A0A232FGJ7 D6WSA1 A0A195C6M8 A0A151XBS1 A0A195EZM6 A0A195EFG6 A0A195BHA1 A0A154PHF5 U4UQT2 E0VNH3 A0A1B6E3W9 A0A0C9PSS7 A0A1B0CDH5 A0A1L8DAX7 A0A067QJS4 A0A1B6J9Y3 A0A1B6ELU9 A0A0A9Y110 B4HJD0 A0A2P8Z2Q5 A0A1I8PNX3 B3P4P6 B4QVX6 W8BGN1 Q9VH60 A0A0K8SQ96 A0A0A1WPR0 Q6NQY0 B4PVG4 A0A1A9X5Y2 A0A1A9V5I5 A0A1B0ASN7 A0A1Q3FVQ7 A0A1Q3FVJ8 A0A1Q3FVJ0 A0A1Q3FVK2 A0A1A9Z745 B4JF41 B4M170 A0A0K8UV86 A0A0P6BQ65 E9HAQ2 A0A0M4ENN6 A0A1W4W070 A0A1I8N9L3 Q299I8 B4G581 A0A0P5PGB4 A0A164YID3 A0A1S4F8W1 A0A0P6ISR2 A0A336LV74 B3M1D3 A0A3B0KCU5 U5EWF2 A0A0P4VN79 B4NKK3 A0A1B0G2I3 A0A0L0CGP8 A0A224X717 A0A069DZL3 A0A023F5L5 B4K952 A0A1J1I2Y4 A0A0P5E1B8 A0A131XXE4 A0A147BWB8 T1HZG2 L7MF40 A0A1E1XB69 A0A224YMR3 A0A3S3NW13 A0A3S4QNS7 T1IYN2 V5H2M0 A0A2S2R020 A0A023GLY2 A0A1E1XQG1 L7MKH5 J9K0E1
PDB
5VMD
E-value=4.20384e-35,
Score=374
Ontologies
GO
PANTHER
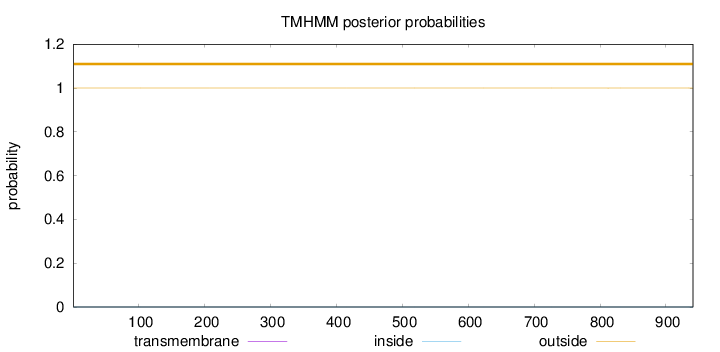
Topology
Length:
941
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00431
Exp number, first 60 AAs:
0.00046
Total prob of N-in:
0.00006
outside
1 - 941
Population Genetic Test Statistics
Pi
252.358231
Theta
189.239514
Tajima's D
0.857479
CLR
0.141499
CSRT
0.621118944052797
Interpretation
Uncertain
