Gene
KWMTBOMO15088
Pre Gene Modal
BGIBMGA014080
Annotation
PREDICTED:_autophagy-related_protein_2_homolog_A-like_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.688 Nuclear Reliability : 1.663
Sequence
CDS
ATGGCCTATATATCGCGCATCCCCCGTGCTGTCATCCGCATAGCAATGCATGCCATCGATAGACAGCATGTCATTGATATACAGGATGAAAAGCGTGGGGGAGAGCACCGAACCTTGTGGAACGCCAGCGTTAATGATCATCGTATAAGAACAGGCACCGTCTACAACGACCGTGATGTTCCGCCCATCCAAAAAGCTAGCGATCCACTTGCAGAGACCCTCGGGGATTCCGTAAGATGGGTCCTGACGTCTCCTGGCATCAACCCATGCCGCGTATGCGGACCGCTTAAGGTGTGCAGCATCCCTACTGACATTGTTATACCAGGGTCTGCCGCGACCCCCAACGGGCACCTCAGAGGAGAGAATAAACAATTCCATCCCCTGGAGCACTACGTCATTAAGACGGTCCGCACAGACGTCAGGATCGGTAGAGGAAAAGCAGAACCGTCCCCATGGGTTAAATCCGGCGGCGATGGGAGCAACACCGACGACGAATACTGTTTCGTGGAGACCCAACCCAGCAGTCAGGACAACGATCTGGATGAGCCAGTCGTGAACTGGATCGGTTCAGGGCCGGCCTACATGTTCGACAATCACTTCAGCGTTCCCGCTGCTAGGACTGATGTACTTAAAGCACCGAAGAGTTTCCCATTGCCGATGTTGCGGTACACGTTGTGCGAAATGAGCATCACATGGAACATGTACGGGGGCAATGACTTCCGTAATCCCGCCGACGCGGTATCTTCCAAGAAAACGGTCACCATAGATGACAATCGTAAATTAGGATCCCCTACCTCTGTAAAGCGCAGTAAAGACTACGAGCCGTACGAGAGCGGCCGCGCCATTGCAGCCGCGTACAGAAGCGGCGGGGTCGGCTGGGCGGCGGGCGCCGACCGAGTGCGGACCAACACGCGCGCGAGCGTGAAGCGCAGGGAACTACGCACCAGGGGCGGACCTCAACGCGACCACAAGACGAAGGTCAAACTCTGCCTCACTAAAGTGAAATTCCAATACGAGGTGTATCCTTCTGGTGGCATGCACGCGTCCCGGCAAACCCTCGCGATCAGCAAGATCGAAGTATTGGATCAACTTGAGTGCAGCGACATCAACAAGCTTCTCAGCCAGTACAAGTTGAAAGACGAGCCGGAGAGAAAAAACGCCCATATGTTAGTTGTGAAAGCAGTACACCTGCGTCCAGACCCCGCACTGACGGCCCAGGAGTGCTGCCTCAAGATATCTCTGTTGCCGCTCTGGTTCAACCTCGACCAAGACACGTTGGGCTTCTTGGTGGGATACTTTTCTAAATTAGGGTCTGATGAATGCAGCGGTGAAGACGACTCAAAAAGCGTCGGAGGTCTGTCGACGGACTCTAGCGGGTCGAGACAGACCACGCCCACCCACCGGCCCCCCGTCATGACCATCGCGAGCCACCTCAAGGACCCGCCCCCCACCCCCACCTCGCTGGGGGACGCTGATGCCCTATCGCTCAACGAAACTGTGCTTCGCGATGAAGAGCCTCTAATGGAGACGTACGAAGCGGAGCGTCTGGTCTCCGAGAACCTGATCCAGCTCGAGGAAGACTTCAACAAACTGGGCATCGATCAAGGGAAACCGGCCGCGAAGCTACCCGTCGACGCTGAGCCCGTCGACGACTCACCGATTTACTTCAGACGCGTGGTGTTTTCCCCGGAAGTACCCATACGATTGGACTACGTGGGCAAACGCGTGGACCTGTCGTCTGGTCCGGTCGCTGGCCTGCTCATGGGACTTGGGCAACTCAACTGCTCCGAGCTCACACTGAAGAGGCTTGACTACAAACTTGGATTGTTGGGCACTGAGAAACTAGTTCAGTGGGCGTTGCACGAATGGCTCTCCGACATAAAGAGGCATCAACTACCGGGATTGCTCAGTGGAATAGGACCGATGCACTCGCTTTTACAACTCATAACCGGCATCCGCGACCTCGTGTGGCTGCCGGTGGAGCAGTGGCGGCGCGACGGGCGGCTGGTGCACGGGCTGCGGCGCGGCGCGGCCTCCTTCACGGCGCGCACTGCCGTCGCCGCGCTCGATATCACCACGCGCATCCTGCACCTCATACAGGCCACAGCAGAGACAGCATTCGACATGCTGACCCCCGGGCCGACCCTGCAGCTGCAAGACGCGTACAACCGCCGGGAGAGGCGTCGCCGGCGGCGGCTGGACCCCTCGCGTCACCCTACCGACATCAGAGACGGAGTGGCCAGCGCCTATCAAACAGTCCGAGAGGGCTTCGCGGAGACAGCGGCGACGGTGAGTCTGGCGGCGCGCGAGTCTAGCGGCGTGGGCGTGCTGCGCATGCTGCCCGGCGCCGCGGTGTCGCCGCTGCTGCTGGCGGCCGCGGGCGCCGCCGACCTGCTGGGCGGGGTGCGCGCCACGCTGGCGCCGCACGAGCGGGGACACTGCGCCGACAAGTGGCGCCGCCACCCTACTCCGGACGCCGTGGACGATTGA
Protein
MAYISRIPRAVIRIAMHAIDRQHVIDIQDEKRGGEHRTLWNASVNDHRIRTGTVYNDRDVPPIQKASDPLAETLGDSVRWVLTSPGINPCRVCGPLKVCSIPTDIVIPGSAATPNGHLRGENKQFHPLEHYVIKTVRTDVRIGRGKAEPSPWVKSGGDGSNTDDEYCFVETQPSSQDNDLDEPVVNWIGSGPAYMFDNHFSVPAARTDVLKAPKSFPLPMLRYTLCEMSITWNMYGGNDFRNPADAVSSKKTVTIDDNRKLGSPTSVKRSKDYEPYESGRAIAAAYRSGGVGWAAGADRVRTNTRASVKRRELRTRGGPQRDHKTKVKLCLTKVKFQYEVYPSGGMHASRQTLAISKIEVLDQLECSDINKLLSQYKLKDEPERKNAHMLVVKAVHLRPDPALTAQECCLKISLLPLWFNLDQDTLGFLVGYFSKLGSDECSGEDDSKSVGGLSTDSSGSRQTTPTHRPPVMTIASHLKDPPPTPTSLGDADALSLNETVLRDEEPLMETYEAERLVSENLIQLEEDFNKLGIDQGKPAAKLPVDAEPVDDSPIYFRRVVFSPEVPIRLDYVGKRVDLSSGPVAGLLMGLGQLNCSELTLKRLDYKLGLLGTEKLVQWALHEWLSDIKRHQLPGLLSGIGPMHSLLQLITGIRDLVWLPVEQWRRDGRLVHGLRRGAASFTARTAVAALDITTRILHLIQATAETAFDMLTPGPTLQLQDAYNRRERRRRRRLDPSRHPTDIRDGVASAYQTVREGFAETAATVSLAARESSGVGVLRMLPGAAVSPLLLAAAGAADLLGGVRATLAPHERGHCADKWRRHPTPDAVDD
Summary
Uniprot
A0A1E1W624
A0A194PFZ7
A0A212ERB3
A0A2H1W423
A0A1Y1M7G5
A0A1Y1M2J6
+ More
A0A139WEE2 A0A182GP91 D6WPD4 A0A182GHW3 A0A0L7R1L6 A0A182M5K2 A0A182L7L8 A0A182RID7 A0A2A3EQF2 A0A088A0E9 Q17EJ4 A0A182V062 A0A182W0F9 A0A1I8JVR9 A0A182Y760 A0A182K4W6 Q7QBB6 A0A2J7PZ47 A0A2C9GPY0 A0A182TCX7 A0A2J7PZ50 A0A336LYP4 A0A182FAT7 W5J3H3 A0A0T6B0W9 B0X651 A0A182PNM0 A0A182QX62 A0A2M3ZZJ7 A0A232EP42 A0A2M3ZZ30 A0A2M4B807 A0A1Q3FXZ6 A0A1Q3FY49 A0A0M8ZVG2 A0A026W559 A0A182N5K0 A0A1Q3FUR3 A0A1Q3FUU4 A0A3L8D4K5 A0A182IY01 A0A084VCJ1 E2BCN5 A0A1L8DVF7 A0A1L8DVE2 A0A154P2Q2 E2ATP3 A0A146KUZ5 A0A0A9Y0W2 U4UK95 N6TWZ4 A0A151JC29 A0A1J1IRZ7 A0A1B6D799 A0A151WUL5 A0A151JWS4 A0A224XCK3 A0A195BX82 A0A158NIS8 F4WDD5 A0A195C9X4 A0A0J7KWF9 A0A2S2R827 E0VNF9 A0A0P6E0Z5 A0A0P6F3J4 A0A0L0C4Y2 A0A164TG66 A0A293N6S4 A0A1B0CUM3 A0A0P5SPR2 A0A0P6I755 A0A0P6IFL7 A0A0P5XG31 A0A0P6B2Q7 T1PF62 A0A1I8MR16 A0A1I8PZM8 A0A226DD79 J9KAE7 A0A3R7SNZ9 A0A2H8TGD4 A0A3P4RNI1
A0A139WEE2 A0A182GP91 D6WPD4 A0A182GHW3 A0A0L7R1L6 A0A182M5K2 A0A182L7L8 A0A182RID7 A0A2A3EQF2 A0A088A0E9 Q17EJ4 A0A182V062 A0A182W0F9 A0A1I8JVR9 A0A182Y760 A0A182K4W6 Q7QBB6 A0A2J7PZ47 A0A2C9GPY0 A0A182TCX7 A0A2J7PZ50 A0A336LYP4 A0A182FAT7 W5J3H3 A0A0T6B0W9 B0X651 A0A182PNM0 A0A182QX62 A0A2M3ZZJ7 A0A232EP42 A0A2M3ZZ30 A0A2M4B807 A0A1Q3FXZ6 A0A1Q3FY49 A0A0M8ZVG2 A0A026W559 A0A182N5K0 A0A1Q3FUR3 A0A1Q3FUU4 A0A3L8D4K5 A0A182IY01 A0A084VCJ1 E2BCN5 A0A1L8DVF7 A0A1L8DVE2 A0A154P2Q2 E2ATP3 A0A146KUZ5 A0A0A9Y0W2 U4UK95 N6TWZ4 A0A151JC29 A0A1J1IRZ7 A0A1B6D799 A0A151WUL5 A0A151JWS4 A0A224XCK3 A0A195BX82 A0A158NIS8 F4WDD5 A0A195C9X4 A0A0J7KWF9 A0A2S2R827 E0VNF9 A0A0P6E0Z5 A0A0P6F3J4 A0A0L0C4Y2 A0A164TG66 A0A293N6S4 A0A1B0CUM3 A0A0P5SPR2 A0A0P6I755 A0A0P6IFL7 A0A0P5XG31 A0A0P6B2Q7 T1PF62 A0A1I8MR16 A0A1I8PZM8 A0A226DD79 J9KAE7 A0A3R7SNZ9 A0A2H8TGD4 A0A3P4RNI1
Pubmed
EMBL
GDQN01008634
JAT82420.1
KQ459604
KPI92217.1
AGBW02013098
OWR43991.1
+ More
ODYU01006127 SOQ47706.1 GEZM01043182 JAV79247.1 GEZM01043181 JAV79248.1 KQ971354 KYB26227.1 JXUM01078091 KQ563044 KXJ74581.1 EFA07398.2 JXUM01064717 KQ562311 KXJ76186.1 KQ414667 KOC64737.1 AXCM01002419 KZ288194 PBC33937.1 CH477282 EAT44885.1 AAAB01008880 EAA08640.5 NEVH01020340 PNF21609.1 APCN01002595 APCN01002596 PNF21608.1 UFQS01000189 UFQT01000189 SSX00997.1 SSX21377.1 ADMH02002196 ETN57873.1 LJIG01016265 KRT81120.1 DS232402 EDS41201.1 AXCN02001163 GGFK01000646 MBW33967.1 NNAY01003012 OXU20123.1 GGFK01000493 MBW33814.1 GGFJ01000043 MBW49184.1 GFDL01002607 JAV32438.1 GFDL01002599 JAV32446.1 KQ435863 KOX70359.1 KK107405 EZA51215.1 GFDL01003706 JAV31339.1 GFDL01003708 JAV31337.1 QOIP01000014 RLU14943.1 ATLV01010765 KE524612 KFB35685.1 GL447336 EFN86545.1 GFDF01003674 JAV10410.1 GFDF01003673 JAV10411.1 KQ434809 KZC06215.1 GL442637 EFN63197.1 GDHC01019672 JAP98956.1 GBHO01018318 JAG25286.1 KB632233 ERL90415.1 APGK01057870 KB741282 ENN70837.1 KQ979079 KYN22683.1 CVRI01000059 CRL03003.1 GEDC01015740 JAS21558.1 KQ982730 KYQ51543.1 KQ981667 KYN38464.1 GFTR01008928 JAW07498.1 KQ976394 KYM93249.1 ADTU01017115 ADTU01017116 GL888087 EGI67715.1 KQ978068 KYM97612.1 LBMM01002400 KMQ94872.1 GGMS01016983 MBY86186.1 DS235339 EEB14915.1 GDIQ01081173 JAN13564.1 GDIQ01068336 JAN26401.1 JRES01000902 KNC27438.1 LRGB01001800 KZS10430.1 GFWV01023164 MAA47891.1 AJWK01029397 AJWK01029398 AJWK01029399 GDIQ01101218 JAL50508.1 GDIQ01034443 JAN60294.1 GDIQ01005267 JAN89470.1 GDIP01073634 JAM30081.1 GDIP01020857 JAM82858.1 KA646800 AFP61429.1 LNIX01000025 OXA42667.1 ABLF02012006 QCYY01002618 ROT68945.1 GFXV01001381 MBW13186.1 CYRY02042655 VCX36440.1
ODYU01006127 SOQ47706.1 GEZM01043182 JAV79247.1 GEZM01043181 JAV79248.1 KQ971354 KYB26227.1 JXUM01078091 KQ563044 KXJ74581.1 EFA07398.2 JXUM01064717 KQ562311 KXJ76186.1 KQ414667 KOC64737.1 AXCM01002419 KZ288194 PBC33937.1 CH477282 EAT44885.1 AAAB01008880 EAA08640.5 NEVH01020340 PNF21609.1 APCN01002595 APCN01002596 PNF21608.1 UFQS01000189 UFQT01000189 SSX00997.1 SSX21377.1 ADMH02002196 ETN57873.1 LJIG01016265 KRT81120.1 DS232402 EDS41201.1 AXCN02001163 GGFK01000646 MBW33967.1 NNAY01003012 OXU20123.1 GGFK01000493 MBW33814.1 GGFJ01000043 MBW49184.1 GFDL01002607 JAV32438.1 GFDL01002599 JAV32446.1 KQ435863 KOX70359.1 KK107405 EZA51215.1 GFDL01003706 JAV31339.1 GFDL01003708 JAV31337.1 QOIP01000014 RLU14943.1 ATLV01010765 KE524612 KFB35685.1 GL447336 EFN86545.1 GFDF01003674 JAV10410.1 GFDF01003673 JAV10411.1 KQ434809 KZC06215.1 GL442637 EFN63197.1 GDHC01019672 JAP98956.1 GBHO01018318 JAG25286.1 KB632233 ERL90415.1 APGK01057870 KB741282 ENN70837.1 KQ979079 KYN22683.1 CVRI01000059 CRL03003.1 GEDC01015740 JAS21558.1 KQ982730 KYQ51543.1 KQ981667 KYN38464.1 GFTR01008928 JAW07498.1 KQ976394 KYM93249.1 ADTU01017115 ADTU01017116 GL888087 EGI67715.1 KQ978068 KYM97612.1 LBMM01002400 KMQ94872.1 GGMS01016983 MBY86186.1 DS235339 EEB14915.1 GDIQ01081173 JAN13564.1 GDIQ01068336 JAN26401.1 JRES01000902 KNC27438.1 LRGB01001800 KZS10430.1 GFWV01023164 MAA47891.1 AJWK01029397 AJWK01029398 AJWK01029399 GDIQ01101218 JAL50508.1 GDIQ01034443 JAN60294.1 GDIQ01005267 JAN89470.1 GDIP01073634 JAM30081.1 GDIP01020857 JAM82858.1 KA646800 AFP61429.1 LNIX01000025 OXA42667.1 ABLF02012006 QCYY01002618 ROT68945.1 GFXV01001381 MBW13186.1 CYRY02042655 VCX36440.1
Proteomes
UP000053268
UP000007151
UP000007266
UP000069940
UP000249989
UP000053825
+ More
UP000075883 UP000075882 UP000075900 UP000242457 UP000005203 UP000008820 UP000075903 UP000075920 UP000076407 UP000076408 UP000075881 UP000007062 UP000235965 UP000075840 UP000075902 UP000069272 UP000000673 UP000002320 UP000075885 UP000075886 UP000215335 UP000053105 UP000053097 UP000075884 UP000279307 UP000075880 UP000030765 UP000008237 UP000076502 UP000000311 UP000030742 UP000019118 UP000078492 UP000183832 UP000075809 UP000078541 UP000078540 UP000005205 UP000007755 UP000078542 UP000036403 UP000009046 UP000037069 UP000076858 UP000092461 UP000095301 UP000095300 UP000198287 UP000007819 UP000283509
UP000075883 UP000075882 UP000075900 UP000242457 UP000005203 UP000008820 UP000075903 UP000075920 UP000076407 UP000076408 UP000075881 UP000007062 UP000235965 UP000075840 UP000075902 UP000069272 UP000000673 UP000002320 UP000075885 UP000075886 UP000215335 UP000053105 UP000053097 UP000075884 UP000279307 UP000075880 UP000030765 UP000008237 UP000076502 UP000000311 UP000030742 UP000019118 UP000078492 UP000183832 UP000075809 UP000078541 UP000078540 UP000005205 UP000007755 UP000078542 UP000036403 UP000009046 UP000037069 UP000076858 UP000092461 UP000095301 UP000095300 UP000198287 UP000007819 UP000283509
Interpro
ProteinModelPortal
A0A1E1W624
A0A194PFZ7
A0A212ERB3
A0A2H1W423
A0A1Y1M7G5
A0A1Y1M2J6
+ More
A0A139WEE2 A0A182GP91 D6WPD4 A0A182GHW3 A0A0L7R1L6 A0A182M5K2 A0A182L7L8 A0A182RID7 A0A2A3EQF2 A0A088A0E9 Q17EJ4 A0A182V062 A0A182W0F9 A0A1I8JVR9 A0A182Y760 A0A182K4W6 Q7QBB6 A0A2J7PZ47 A0A2C9GPY0 A0A182TCX7 A0A2J7PZ50 A0A336LYP4 A0A182FAT7 W5J3H3 A0A0T6B0W9 B0X651 A0A182PNM0 A0A182QX62 A0A2M3ZZJ7 A0A232EP42 A0A2M3ZZ30 A0A2M4B807 A0A1Q3FXZ6 A0A1Q3FY49 A0A0M8ZVG2 A0A026W559 A0A182N5K0 A0A1Q3FUR3 A0A1Q3FUU4 A0A3L8D4K5 A0A182IY01 A0A084VCJ1 E2BCN5 A0A1L8DVF7 A0A1L8DVE2 A0A154P2Q2 E2ATP3 A0A146KUZ5 A0A0A9Y0W2 U4UK95 N6TWZ4 A0A151JC29 A0A1J1IRZ7 A0A1B6D799 A0A151WUL5 A0A151JWS4 A0A224XCK3 A0A195BX82 A0A158NIS8 F4WDD5 A0A195C9X4 A0A0J7KWF9 A0A2S2R827 E0VNF9 A0A0P6E0Z5 A0A0P6F3J4 A0A0L0C4Y2 A0A164TG66 A0A293N6S4 A0A1B0CUM3 A0A0P5SPR2 A0A0P6I755 A0A0P6IFL7 A0A0P5XG31 A0A0P6B2Q7 T1PF62 A0A1I8MR16 A0A1I8PZM8 A0A226DD79 J9KAE7 A0A3R7SNZ9 A0A2H8TGD4 A0A3P4RNI1
A0A139WEE2 A0A182GP91 D6WPD4 A0A182GHW3 A0A0L7R1L6 A0A182M5K2 A0A182L7L8 A0A182RID7 A0A2A3EQF2 A0A088A0E9 Q17EJ4 A0A182V062 A0A182W0F9 A0A1I8JVR9 A0A182Y760 A0A182K4W6 Q7QBB6 A0A2J7PZ47 A0A2C9GPY0 A0A182TCX7 A0A2J7PZ50 A0A336LYP4 A0A182FAT7 W5J3H3 A0A0T6B0W9 B0X651 A0A182PNM0 A0A182QX62 A0A2M3ZZJ7 A0A232EP42 A0A2M3ZZ30 A0A2M4B807 A0A1Q3FXZ6 A0A1Q3FY49 A0A0M8ZVG2 A0A026W559 A0A182N5K0 A0A1Q3FUR3 A0A1Q3FUU4 A0A3L8D4K5 A0A182IY01 A0A084VCJ1 E2BCN5 A0A1L8DVF7 A0A1L8DVE2 A0A154P2Q2 E2ATP3 A0A146KUZ5 A0A0A9Y0W2 U4UK95 N6TWZ4 A0A151JC29 A0A1J1IRZ7 A0A1B6D799 A0A151WUL5 A0A151JWS4 A0A224XCK3 A0A195BX82 A0A158NIS8 F4WDD5 A0A195C9X4 A0A0J7KWF9 A0A2S2R827 E0VNF9 A0A0P6E0Z5 A0A0P6F3J4 A0A0L0C4Y2 A0A164TG66 A0A293N6S4 A0A1B0CUM3 A0A0P5SPR2 A0A0P6I755 A0A0P6IFL7 A0A0P5XG31 A0A0P6B2Q7 T1PF62 A0A1I8MR16 A0A1I8PZM8 A0A226DD79 J9KAE7 A0A3R7SNZ9 A0A2H8TGD4 A0A3P4RNI1
Ontologies
KEGG
PANTHER
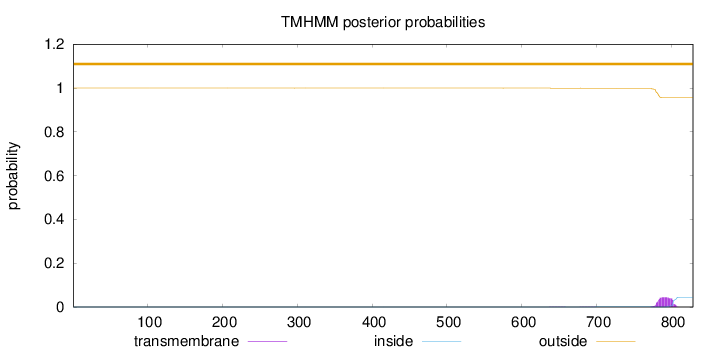
Topology
Length:
829
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.98174
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00020
outside
1 - 829
Population Genetic Test Statistics
Pi
3.743288
Theta
171.464621
Tajima's D
1.339427
CLR
0.294404
CSRT
0.749862506874656
Interpretation
Uncertain
