Pre Gene Modal
BGIBMGA004839
Annotation
PREDICTED:_39S_ribosomal_protein_L40?_mitochondrial_[Plutella_xylostella]
Location in the cell
Nuclear Reliability : 3.231
Sequence
CDS
ATGAAGAAAAAAAAGAAAATAGATCCTGCTATTGTTAAAGCCCGTGAAGAGCGCCGTCGTAAGAAAATTGAGAAACAAATTCGAAGGCTTGAGAAGAATGCTAGACAGTTGAAACCTATTGATGAGTTGGAGGTTCCACTTCACTTGATGGATTCACTCAAAAAATATAAACGGCCTCCTGTTCAGTTGTCGGTCGAAGAAATCGAAGCCCGCGAGCTACTCCAAAAGGAATGGGCTCGTTACAAACGTGATGAATATATGAACAACATTGCACAAGTCGACAGGATCATGGCAGCCCAGAGACGCGCTCTGGACAGGCTGTACGAGGAATCAGAGGATCTGTACAATGAAGCTATTATGCCCGACTTGCAACTACTGCCTTATACCATAAGCGGTCCGGTAGCGACGCCGCCCATCAAGGACTACGAATCGCCCGACGGAGAATACATTGACGTGTCGAAGAAATGGGATAATTAG
Protein
MKKKKKIDPAIVKAREERRRKKIEKQIRRLEKNARQLKPIDELEVPLHLMDSLKKYKRPPVQLSVEEIEARELLQKEWARYKRDEYMNNIAQVDRIMAAQRRALDRLYEESEDLYNEAIMPDLQLLPYTISGPVATPPIKDYESPDGEYIDVSKKWDN
Summary
Similarity
Belongs to the cyclin family.
Uniprot
H9J5P9
I4DKY1
A0A2A4J3L2
A0A2H1WZA9
A0A194RKC1
A0A212F2Y0
+ More
A0A194PGP8 U5EFX2 A0A0K8TSB5 A0A1B0CY61 A0A084WR16 A0A0A1X823 A0A1L8DRS8 A0A1L8DRA6 A0A182QWZ8 A0A084WFK5 A0A0L0C4A7 A0A182W413 A0A2P8XMM4 A0A182PLM9 A0A0C9R2U3 A0A182RVS9 A0A182LYW2 A0A3B0JNM1 A0A182YQW4 A0A182JUW4 A0A026WQ92 E1ZXL6 E2BLK1 A0A182N049 A0A182VK94 A0A2J7R4T4 B4NAY0 Q170X2 A0A1I8PID9 W5J1N0 V5GR12 T1EAH3 A0A182HIJ9 A0A182H0Q5 A0A182IS59 A0A336L344 A0A182KZA3 Q7QER3 D6WU16 A0A023EJE3 A0A0M3QXR2 A0A0T6AU57 A0A1S4FHQ6 A0A336M717 A0A182UC70 A0A1W4XBW2 A0A1L8EGU6 A0A1L8EGQ3 A0A1Y1NL65 A0A182WY70 N6UHL5 Q9VGP7 J3JWE9 A0A2M3ZJH1 A0A195F2T7 A0A2M4AV29 A0A2M4AV23 A0A067R3P4 U4U168 A0A2M4C0Y1 B4HIA1 A0A1A9V4P7 A0A1W4USY0 B4PM48 A0A0J7L1R7 W8BJ38 A0A1A9WT43 F4X3T7 A0A1B0G2N9 B4QU45 A0A182T3M0 A0A0A9YP58 A0A1A9XHK3 A0A0K8SNV1 B4K9U2 B3NZF7 B3LXD0 A0A1Q3FBL9 A0A1B6KI01 A0A1B6JCZ3 A0A1W4X2F0 A0A226EL63 A0A182FHU4 B0WN54 E9ISY7 A0A1B6DHD1 A0A1A9ZLX7 A0A151XJC0 A0A1B6F113 A0A1I8MXJ5 A0A2S2Q9T0 A0A195BGL8 Q293S8
A0A194PGP8 U5EFX2 A0A0K8TSB5 A0A1B0CY61 A0A084WR16 A0A0A1X823 A0A1L8DRS8 A0A1L8DRA6 A0A182QWZ8 A0A084WFK5 A0A0L0C4A7 A0A182W413 A0A2P8XMM4 A0A182PLM9 A0A0C9R2U3 A0A182RVS9 A0A182LYW2 A0A3B0JNM1 A0A182YQW4 A0A182JUW4 A0A026WQ92 E1ZXL6 E2BLK1 A0A182N049 A0A182VK94 A0A2J7R4T4 B4NAY0 Q170X2 A0A1I8PID9 W5J1N0 V5GR12 T1EAH3 A0A182HIJ9 A0A182H0Q5 A0A182IS59 A0A336L344 A0A182KZA3 Q7QER3 D6WU16 A0A023EJE3 A0A0M3QXR2 A0A0T6AU57 A0A1S4FHQ6 A0A336M717 A0A182UC70 A0A1W4XBW2 A0A1L8EGU6 A0A1L8EGQ3 A0A1Y1NL65 A0A182WY70 N6UHL5 Q9VGP7 J3JWE9 A0A2M3ZJH1 A0A195F2T7 A0A2M4AV29 A0A2M4AV23 A0A067R3P4 U4U168 A0A2M4C0Y1 B4HIA1 A0A1A9V4P7 A0A1W4USY0 B4PM48 A0A0J7L1R7 W8BJ38 A0A1A9WT43 F4X3T7 A0A1B0G2N9 B4QU45 A0A182T3M0 A0A0A9YP58 A0A1A9XHK3 A0A0K8SNV1 B4K9U2 B3NZF7 B3LXD0 A0A1Q3FBL9 A0A1B6KI01 A0A1B6JCZ3 A0A1W4X2F0 A0A226EL63 A0A182FHU4 B0WN54 E9ISY7 A0A1B6DHD1 A0A1A9ZLX7 A0A151XJC0 A0A1B6F113 A0A1I8MXJ5 A0A2S2Q9T0 A0A195BGL8 Q293S8
Pubmed
19121390
22651552
26354079
22118469
26369729
24438588
+ More
25830018 26108605 29403074 25244985 24508170 30249741 20798317 17994087 17510324 20920257 23761445 26483478 20966253 12364791 14747013 17210077 18362917 19820115 24945155 28004739 23537049 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22516182 24845553 17550304 24495485 21719571 25401762 26823975 21282665 25315136 15632085
25830018 26108605 29403074 25244985 24508170 30249741 20798317 17994087 17510324 20920257 23761445 26483478 20966253 12364791 14747013 17210077 18362917 19820115 24945155 28004739 23537049 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22516182 24845553 17550304 24495485 21719571 25401762 26823975 21282665 25315136 15632085
EMBL
BABH01024961
AK401949
BAM18571.1
NWSH01003221
PCG66737.1
ODYU01012170
+ More
SOQ58342.1 KQ460045 KPJ18278.1 AGBW02010665 OWR48084.1 KQ459604 KPI92213.1 GANO01003698 JAB56173.1 GDAI01000572 JAI17031.1 AJWK01035490 ATLV01025887 KE525402 KFB52660.1 GBXI01007417 JAD06875.1 GFDF01005049 JAV09035.1 GFDF01005143 JAV08941.1 AXCN02002023 ATLV01023372 KE525342 KFB48999.1 JRES01000936 KNC27067.1 PYGN01001711 PSN33246.1 GBYB01010529 JAG80296.1 AXCM01003165 OUUW01000008 SPP83834.1 KK107139 QOIP01000003 EZA57866.1 RLU24667.1 GL435066 EFN74099.1 GL449033 EFN83438.1 NEVH01007402 PNF35846.1 CH964232 EDW80944.1 CH477463 EAT40537.1 ADMH02002165 ETN58122.1 GALX01001882 JAB66584.1 GAMD01000412 JAB01179.1 APCN01003991 JXUM01101760 KQ564673 KXJ71920.1 UFQS01001426 UFQT01001426 SSX10823.1 SSX30503.1 AAAB01008846 EAA06344.3 KQ971352 EFA07359.2 GAPW01004528 JAC09070.1 CP012526 ALC46373.1 LJIG01022834 KRT78477.1 UFQT01000550 SSX25181.1 GFDG01000888 JAV17911.1 GFDG01000887 JAV17912.1 GEZM01000679 JAV98318.1 APGK01025660 KB740600 ENN80091.1 AE014297 AY071609 AAF54629.1 AAL49231.1 BT127567 AEE62529.1 GGFM01007986 MBW28737.1 KQ981856 KYN34492.1 GGFK01011300 MBW44621.1 GGFK01011299 MBW44620.1 KK852718 KDR17788.1 KB631982 ERL87614.1 GGFJ01009690 MBW58831.1 CH480815 EDW42616.1 CM000160 EDW96914.1 LBMM01001273 KMQ96606.1 GAMC01013264 JAB93291.1 GL888624 EGI58884.1 CCAG010000815 CM000364 EDX13376.1 GBHO01008777 GBHO01008776 JAG34827.1 JAG34828.1 GBRD01010906 GDHC01012367 JAG54918.1 JAQ06262.1 CH933806 EDW15590.1 CH954181 EDV49530.1 CH902617 EDV42774.1 GFDL01010136 JAV24909.1 GEBQ01029193 JAT10784.1 GECU01010644 JAS97062.1 LNIX01000003 OXA58199.1 DS232007 EDS31485.1 GL765434 EFZ16308.1 GEDC01012187 JAS25111.1 KQ982074 KYQ60516.1 GECZ01025925 JAS43844.1 GGMS01005306 MBY74509.1 KQ976490 KYM83366.1 CM000070 EAL29136.2
SOQ58342.1 KQ460045 KPJ18278.1 AGBW02010665 OWR48084.1 KQ459604 KPI92213.1 GANO01003698 JAB56173.1 GDAI01000572 JAI17031.1 AJWK01035490 ATLV01025887 KE525402 KFB52660.1 GBXI01007417 JAD06875.1 GFDF01005049 JAV09035.1 GFDF01005143 JAV08941.1 AXCN02002023 ATLV01023372 KE525342 KFB48999.1 JRES01000936 KNC27067.1 PYGN01001711 PSN33246.1 GBYB01010529 JAG80296.1 AXCM01003165 OUUW01000008 SPP83834.1 KK107139 QOIP01000003 EZA57866.1 RLU24667.1 GL435066 EFN74099.1 GL449033 EFN83438.1 NEVH01007402 PNF35846.1 CH964232 EDW80944.1 CH477463 EAT40537.1 ADMH02002165 ETN58122.1 GALX01001882 JAB66584.1 GAMD01000412 JAB01179.1 APCN01003991 JXUM01101760 KQ564673 KXJ71920.1 UFQS01001426 UFQT01001426 SSX10823.1 SSX30503.1 AAAB01008846 EAA06344.3 KQ971352 EFA07359.2 GAPW01004528 JAC09070.1 CP012526 ALC46373.1 LJIG01022834 KRT78477.1 UFQT01000550 SSX25181.1 GFDG01000888 JAV17911.1 GFDG01000887 JAV17912.1 GEZM01000679 JAV98318.1 APGK01025660 KB740600 ENN80091.1 AE014297 AY071609 AAF54629.1 AAL49231.1 BT127567 AEE62529.1 GGFM01007986 MBW28737.1 KQ981856 KYN34492.1 GGFK01011300 MBW44621.1 GGFK01011299 MBW44620.1 KK852718 KDR17788.1 KB631982 ERL87614.1 GGFJ01009690 MBW58831.1 CH480815 EDW42616.1 CM000160 EDW96914.1 LBMM01001273 KMQ96606.1 GAMC01013264 JAB93291.1 GL888624 EGI58884.1 CCAG010000815 CM000364 EDX13376.1 GBHO01008777 GBHO01008776 JAG34827.1 JAG34828.1 GBRD01010906 GDHC01012367 JAG54918.1 JAQ06262.1 CH933806 EDW15590.1 CH954181 EDV49530.1 CH902617 EDV42774.1 GFDL01010136 JAV24909.1 GEBQ01029193 JAT10784.1 GECU01010644 JAS97062.1 LNIX01000003 OXA58199.1 DS232007 EDS31485.1 GL765434 EFZ16308.1 GEDC01012187 JAS25111.1 KQ982074 KYQ60516.1 GECZ01025925 JAS43844.1 GGMS01005306 MBY74509.1 KQ976490 KYM83366.1 CM000070 EAL29136.2
Proteomes
UP000005204
UP000218220
UP000053240
UP000007151
UP000053268
UP000092461
+ More
UP000030765 UP000075886 UP000037069 UP000075920 UP000245037 UP000075885 UP000075900 UP000075883 UP000268350 UP000076408 UP000075881 UP000053097 UP000279307 UP000000311 UP000008237 UP000075884 UP000075903 UP000235965 UP000007798 UP000008820 UP000095300 UP000000673 UP000075840 UP000069940 UP000249989 UP000075880 UP000075882 UP000007062 UP000007266 UP000092553 UP000075902 UP000192223 UP000076407 UP000019118 UP000000803 UP000078541 UP000027135 UP000030742 UP000001292 UP000078200 UP000192221 UP000002282 UP000036403 UP000091820 UP000007755 UP000092444 UP000000304 UP000075901 UP000092443 UP000009192 UP000008711 UP000007801 UP000198287 UP000069272 UP000002320 UP000092445 UP000075809 UP000095301 UP000078540 UP000001819
UP000030765 UP000075886 UP000037069 UP000075920 UP000245037 UP000075885 UP000075900 UP000075883 UP000268350 UP000076408 UP000075881 UP000053097 UP000279307 UP000000311 UP000008237 UP000075884 UP000075903 UP000235965 UP000007798 UP000008820 UP000095300 UP000000673 UP000075840 UP000069940 UP000249989 UP000075880 UP000075882 UP000007062 UP000007266 UP000092553 UP000075902 UP000192223 UP000076407 UP000019118 UP000000803 UP000078541 UP000027135 UP000030742 UP000001292 UP000078200 UP000192221 UP000002282 UP000036403 UP000091820 UP000007755 UP000092444 UP000000304 UP000075901 UP000092443 UP000009192 UP000008711 UP000007801 UP000198287 UP000069272 UP000002320 UP000092445 UP000075809 UP000095301 UP000078540 UP000001819
Interpro
SUPFAM
SSF47954
SSF47954
CDD
ProteinModelPortal
H9J5P9
I4DKY1
A0A2A4J3L2
A0A2H1WZA9
A0A194RKC1
A0A212F2Y0
+ More
A0A194PGP8 U5EFX2 A0A0K8TSB5 A0A1B0CY61 A0A084WR16 A0A0A1X823 A0A1L8DRS8 A0A1L8DRA6 A0A182QWZ8 A0A084WFK5 A0A0L0C4A7 A0A182W413 A0A2P8XMM4 A0A182PLM9 A0A0C9R2U3 A0A182RVS9 A0A182LYW2 A0A3B0JNM1 A0A182YQW4 A0A182JUW4 A0A026WQ92 E1ZXL6 E2BLK1 A0A182N049 A0A182VK94 A0A2J7R4T4 B4NAY0 Q170X2 A0A1I8PID9 W5J1N0 V5GR12 T1EAH3 A0A182HIJ9 A0A182H0Q5 A0A182IS59 A0A336L344 A0A182KZA3 Q7QER3 D6WU16 A0A023EJE3 A0A0M3QXR2 A0A0T6AU57 A0A1S4FHQ6 A0A336M717 A0A182UC70 A0A1W4XBW2 A0A1L8EGU6 A0A1L8EGQ3 A0A1Y1NL65 A0A182WY70 N6UHL5 Q9VGP7 J3JWE9 A0A2M3ZJH1 A0A195F2T7 A0A2M4AV29 A0A2M4AV23 A0A067R3P4 U4U168 A0A2M4C0Y1 B4HIA1 A0A1A9V4P7 A0A1W4USY0 B4PM48 A0A0J7L1R7 W8BJ38 A0A1A9WT43 F4X3T7 A0A1B0G2N9 B4QU45 A0A182T3M0 A0A0A9YP58 A0A1A9XHK3 A0A0K8SNV1 B4K9U2 B3NZF7 B3LXD0 A0A1Q3FBL9 A0A1B6KI01 A0A1B6JCZ3 A0A1W4X2F0 A0A226EL63 A0A182FHU4 B0WN54 E9ISY7 A0A1B6DHD1 A0A1A9ZLX7 A0A151XJC0 A0A1B6F113 A0A1I8MXJ5 A0A2S2Q9T0 A0A195BGL8 Q293S8
A0A194PGP8 U5EFX2 A0A0K8TSB5 A0A1B0CY61 A0A084WR16 A0A0A1X823 A0A1L8DRS8 A0A1L8DRA6 A0A182QWZ8 A0A084WFK5 A0A0L0C4A7 A0A182W413 A0A2P8XMM4 A0A182PLM9 A0A0C9R2U3 A0A182RVS9 A0A182LYW2 A0A3B0JNM1 A0A182YQW4 A0A182JUW4 A0A026WQ92 E1ZXL6 E2BLK1 A0A182N049 A0A182VK94 A0A2J7R4T4 B4NAY0 Q170X2 A0A1I8PID9 W5J1N0 V5GR12 T1EAH3 A0A182HIJ9 A0A182H0Q5 A0A182IS59 A0A336L344 A0A182KZA3 Q7QER3 D6WU16 A0A023EJE3 A0A0M3QXR2 A0A0T6AU57 A0A1S4FHQ6 A0A336M717 A0A182UC70 A0A1W4XBW2 A0A1L8EGU6 A0A1L8EGQ3 A0A1Y1NL65 A0A182WY70 N6UHL5 Q9VGP7 J3JWE9 A0A2M3ZJH1 A0A195F2T7 A0A2M4AV29 A0A2M4AV23 A0A067R3P4 U4U168 A0A2M4C0Y1 B4HIA1 A0A1A9V4P7 A0A1W4USY0 B4PM48 A0A0J7L1R7 W8BJ38 A0A1A9WT43 F4X3T7 A0A1B0G2N9 B4QU45 A0A182T3M0 A0A0A9YP58 A0A1A9XHK3 A0A0K8SNV1 B4K9U2 B3NZF7 B3LXD0 A0A1Q3FBL9 A0A1B6KI01 A0A1B6JCZ3 A0A1W4X2F0 A0A226EL63 A0A182FHU4 B0WN54 E9ISY7 A0A1B6DHD1 A0A1A9ZLX7 A0A151XJC0 A0A1B6F113 A0A1I8MXJ5 A0A2S2Q9T0 A0A195BGL8 Q293S8
PDB
6NU3
E-value=6.81945e-15,
Score=191
Ontologies
PANTHER
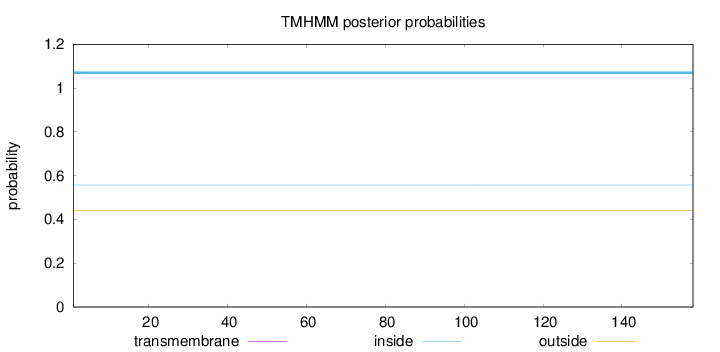
Topology
Length:
158
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00097
Exp number, first 60 AAs:
0
Total prob of N-in:
0.55771
inside
1 - 158
Population Genetic Test Statistics
Pi
318.474762
Theta
192.050333
Tajima's D
2.57099
CLR
0.107627
CSRT
0.948652567371631
Interpretation
Uncertain
