Gene
KWMTBOMO15079
Pre Gene Modal
BGIBMGA004841
Annotation
PREDICTED:_uncharacterized_protein_LOC101741141_isoform_X3_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.505
Sequence
CDS
ATGCGGCGCCTCCTCGTGACGCTGATAGCCATCGCCGCCGACATCTACGCTCAAGACTCGCCCAGGGAACATCAGGGACCACAGATTTGCGGGGGCCGGCTCCGAGGACCCACGGGCGTGATACAGACACCCAATTTCCCCAACCCGTTTCCCGTGCCGATAAAGTGCCGTTGGATTATCGAGCACGACATCGCGAACGGAACTATTTCGATATATTTCACTCAACAATACACGACTAGTGGATTAACCTTCACTGAATACATGTATTATGATGAATCGTATAAGCTGGGCGAGAGGCGGGCTCTAACTGTGACCGAGGACAGCATCACAAGAGTTAAATGGCTGCAGGTAAGCTTATGA
Protein
MRRLLVTLIAIAADIYAQDSPREHQGPQICGGRLRGPTGVIQTPNFPNPFPVPIKCRWIIEHDIANGTISIYFTQQYTTSGLTFTEYMYYDESYKLGERRALTVTEDSITRVKWLQVSL
Summary
Uniprot
H9J5Q1
A0A0L7L2F1
A0A194RQH7
A0A2A4JRC1
A0A2H1X184
A0A194PFQ5
+ More
A0A2W1BIR1 A0A1A9WKE3 A0A182JB14 A0A1B0DAM9 A0A1B0GA86 A0A0L0CG37 A0A182GYS3 Q16H97 Q16ZC4 A0A1B0AJS1 A0A1A9UPM5 A0A1A9XN51 A0A1B0AT93 A0A182RW89 A0A084WG67 A0A1I8PU15 A0A182WGN2 A0A182YDF5 A0A182SZP6 A0A182XGA9 Q7Q1G8 A0A182HRT0 A0A182JY43 B0WGS9 A0A182UFP0 A0A182MN39 A0A182V1X9 A0A1I8M3C5 A0A182KTX1 B4G7L8 A0A182PCU2 A0A154PRY8 A0A2R7WQP9 Q29NG8 A0A182NBV7 A0A3B0JU17 B3N920 A0A1J1I3T9 A0A182QIC5 A0A2A3ELN7 B3MPK9 A0A0P8XGM3 A0A0P8Y4H3 E1ZWL7 A0A088A1Y9 W8BT52 W5JFA1 A0A1W4UFM5 A0A0R1DLB3 T1HCX1 A0A195F423 B4Q8P5 A0A195D9U6 A0A195CEW3 A0A0J7KUZ6 A0A0L7QUB9 E2BQD5 B4HWG3 A0A151XEB2 A0A182F2Y7 A0A2P8Z8P5 A0A195BP00 A0A139WC50 B4JCJ1 F4W506 A0A1B6ILL7 A0A0J9QZQ8 A0A158NVD3 Q9VL42 B4KH51 E9J311 A0A1B6CZL9 A0A0M9AA20 A0A0A9VU98 A0A146LK95 A0A3L8DNQ9 B4NLH4 B4M9D6 A0A2R7WPD6 K7IX39 A0A026X531 T1HCX0 E0V9X9 A0A2P6KAI8 R7VIL3
A0A2W1BIR1 A0A1A9WKE3 A0A182JB14 A0A1B0DAM9 A0A1B0GA86 A0A0L0CG37 A0A182GYS3 Q16H97 Q16ZC4 A0A1B0AJS1 A0A1A9UPM5 A0A1A9XN51 A0A1B0AT93 A0A182RW89 A0A084WG67 A0A1I8PU15 A0A182WGN2 A0A182YDF5 A0A182SZP6 A0A182XGA9 Q7Q1G8 A0A182HRT0 A0A182JY43 B0WGS9 A0A182UFP0 A0A182MN39 A0A182V1X9 A0A1I8M3C5 A0A182KTX1 B4G7L8 A0A182PCU2 A0A154PRY8 A0A2R7WQP9 Q29NG8 A0A182NBV7 A0A3B0JU17 B3N920 A0A1J1I3T9 A0A182QIC5 A0A2A3ELN7 B3MPK9 A0A0P8XGM3 A0A0P8Y4H3 E1ZWL7 A0A088A1Y9 W8BT52 W5JFA1 A0A1W4UFM5 A0A0R1DLB3 T1HCX1 A0A195F423 B4Q8P5 A0A195D9U6 A0A195CEW3 A0A0J7KUZ6 A0A0L7QUB9 E2BQD5 B4HWG3 A0A151XEB2 A0A182F2Y7 A0A2P8Z8P5 A0A195BP00 A0A139WC50 B4JCJ1 F4W506 A0A1B6ILL7 A0A0J9QZQ8 A0A158NVD3 Q9VL42 B4KH51 E9J311 A0A1B6CZL9 A0A0M9AA20 A0A0A9VU98 A0A146LK95 A0A3L8DNQ9 B4NLH4 B4M9D6 A0A2R7WPD6 K7IX39 A0A026X531 T1HCX0 E0V9X9 A0A2P6KAI8 R7VIL3
Pubmed
19121390
26227816
26354079
28756777
26108605
26483478
+ More
17510324 24438588 25244985 12364791 25315136 20966253 17994087 15632085 20798317 24495485 20920257 23761445 17550304 29403074 18362917 19820115 21719571 22936249 21347285 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 21282665 25401762 26823975 30249741 18057021 20075255 24508170 20566863 23254933
17510324 24438588 25244985 12364791 25315136 20966253 17994087 15632085 20798317 24495485 20920257 23761445 17550304 29403074 18362917 19820115 21719571 22936249 21347285 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 21282665 25401762 26823975 30249741 18057021 20075255 24508170 20566863 23254933
EMBL
BABH01024992
JTDY01003498
KOB69464.1
KQ460045
KPJ18281.1
NWSH01000768
+ More
PCG74316.1 ODYU01012650 SOQ59079.1 KQ459604 KPI92211.1 KZ150294 PZC71563.1 AJVK01004526 CCAG010019844 CCAG010019845 CCAG010019846 CCAG010019847 JRES01000523 KNC30439.1 JXUM01098114 KQ564395 KXJ72342.1 CH478189 EAT33619.1 CH477493 EAT40003.1 JXJN01003205 JXJN01003206 ATLV01023453 KE525343 KFB49211.1 AAAB01008980 EAA14277.3 APCN01001743 DS231929 EDS27184.1 AXCM01002313 CH479180 EDW28418.1 KQ435078 KZC14507.1 KK855299 PTY21894.1 CH379060 EAL33374.3 OUUW01000010 SPP85607.1 CH954177 EDV58455.2 CVRI01000036 CRK93033.1 AXCN02000691 KZ288215 PBC32638.1 CH902620 EDV32257.2 KPU73861.1 KPU73862.1 GL434853 EFN74454.1 GAMC01002030 JAC04526.1 ADMH02001647 ETN61560.1 CM000157 KRJ97992.1 ACPB03000346 KQ981820 KYN35213.1 CM000361 EDX04460.1 KQ981082 KYN09641.1 KQ977827 KYM99429.1 LBMM01002830 KMQ94307.1 KQ414735 KOC62238.1 GL449738 EFN82099.1 CH480818 EDW52358.1 KQ982254 KYQ58716.1 PYGN01000145 PSN52875.1 KQ976424 KYM88240.1 KQ971372 KYB25425.1 CH916368 EDW03145.1 GL887596 EGI70702.1 GECU01019893 JAS87813.1 CM002910 KMY89398.1 ADTU01027093 AE014134 AAF52856.4 AGB92849.1 CH933807 EDW12262.2 GL768055 EFZ12772.1 GEDC01018408 JAS18890.1 KQ435716 KOX79023.1 GBHO01043757 JAF99846.1 GDHC01010115 JAQ08514.1 QOIP01000006 RLU21863.1 CH964274 EDW85213.2 CH940654 EDW57812.2 KRF77960.1 KK855217 PTY21498.1 AAZX01000615 KK107020 EZA62539.1 ACPB03000334 ACPB03000335 DS235000 EEB10185.1 MWRG01017965 PRD23338.1 AMQN01004347 KB293301 ELU16131.1
PCG74316.1 ODYU01012650 SOQ59079.1 KQ459604 KPI92211.1 KZ150294 PZC71563.1 AJVK01004526 CCAG010019844 CCAG010019845 CCAG010019846 CCAG010019847 JRES01000523 KNC30439.1 JXUM01098114 KQ564395 KXJ72342.1 CH478189 EAT33619.1 CH477493 EAT40003.1 JXJN01003205 JXJN01003206 ATLV01023453 KE525343 KFB49211.1 AAAB01008980 EAA14277.3 APCN01001743 DS231929 EDS27184.1 AXCM01002313 CH479180 EDW28418.1 KQ435078 KZC14507.1 KK855299 PTY21894.1 CH379060 EAL33374.3 OUUW01000010 SPP85607.1 CH954177 EDV58455.2 CVRI01000036 CRK93033.1 AXCN02000691 KZ288215 PBC32638.1 CH902620 EDV32257.2 KPU73861.1 KPU73862.1 GL434853 EFN74454.1 GAMC01002030 JAC04526.1 ADMH02001647 ETN61560.1 CM000157 KRJ97992.1 ACPB03000346 KQ981820 KYN35213.1 CM000361 EDX04460.1 KQ981082 KYN09641.1 KQ977827 KYM99429.1 LBMM01002830 KMQ94307.1 KQ414735 KOC62238.1 GL449738 EFN82099.1 CH480818 EDW52358.1 KQ982254 KYQ58716.1 PYGN01000145 PSN52875.1 KQ976424 KYM88240.1 KQ971372 KYB25425.1 CH916368 EDW03145.1 GL887596 EGI70702.1 GECU01019893 JAS87813.1 CM002910 KMY89398.1 ADTU01027093 AE014134 AAF52856.4 AGB92849.1 CH933807 EDW12262.2 GL768055 EFZ12772.1 GEDC01018408 JAS18890.1 KQ435716 KOX79023.1 GBHO01043757 JAF99846.1 GDHC01010115 JAQ08514.1 QOIP01000006 RLU21863.1 CH964274 EDW85213.2 CH940654 EDW57812.2 KRF77960.1 KK855217 PTY21498.1 AAZX01000615 KK107020 EZA62539.1 ACPB03000334 ACPB03000335 DS235000 EEB10185.1 MWRG01017965 PRD23338.1 AMQN01004347 KB293301 ELU16131.1
Proteomes
UP000005204
UP000037510
UP000053240
UP000218220
UP000053268
UP000091820
+ More
UP000075880 UP000092462 UP000092444 UP000037069 UP000069940 UP000249989 UP000008820 UP000092445 UP000078200 UP000092443 UP000092460 UP000075900 UP000030765 UP000095300 UP000075920 UP000076408 UP000075901 UP000076407 UP000007062 UP000075840 UP000075881 UP000002320 UP000075902 UP000075883 UP000075903 UP000095301 UP000075882 UP000008744 UP000075885 UP000076502 UP000001819 UP000075884 UP000268350 UP000008711 UP000183832 UP000075886 UP000242457 UP000007801 UP000000311 UP000005203 UP000000673 UP000192221 UP000002282 UP000015103 UP000078541 UP000000304 UP000078492 UP000078542 UP000036403 UP000053825 UP000008237 UP000001292 UP000075809 UP000069272 UP000245037 UP000078540 UP000007266 UP000001070 UP000007755 UP000005205 UP000000803 UP000009192 UP000053105 UP000279307 UP000007798 UP000008792 UP000002358 UP000053097 UP000009046 UP000014760
UP000075880 UP000092462 UP000092444 UP000037069 UP000069940 UP000249989 UP000008820 UP000092445 UP000078200 UP000092443 UP000092460 UP000075900 UP000030765 UP000095300 UP000075920 UP000076408 UP000075901 UP000076407 UP000007062 UP000075840 UP000075881 UP000002320 UP000075902 UP000075883 UP000075903 UP000095301 UP000075882 UP000008744 UP000075885 UP000076502 UP000001819 UP000075884 UP000268350 UP000008711 UP000183832 UP000075886 UP000242457 UP000007801 UP000000311 UP000005203 UP000000673 UP000192221 UP000002282 UP000015103 UP000078541 UP000000304 UP000078492 UP000078542 UP000036403 UP000053825 UP000008237 UP000001292 UP000075809 UP000069272 UP000245037 UP000078540 UP000007266 UP000001070 UP000007755 UP000005205 UP000000803 UP000009192 UP000053105 UP000279307 UP000007798 UP000008792 UP000002358 UP000053097 UP000009046 UP000014760
Interpro
IPR035914
Sperma_CUB_dom_sf
+ More
IPR000859 CUB_dom
IPR036445 GPCR_2_extracell_dom_sf
IPR013032 EGF-like_CS
IPR000742 EGF-like_dom
IPR036179 Ig-like_dom_sf
IPR001879 GPCR_2_extracellular_dom
IPR003599 Ig_sub
IPR007110 Ig-like_dom
IPR013783 Ig-like_fold
IPR003598 Ig_sub2
IPR000203 GPS
IPR029064 L30e-like
IPR001881 EGF-like_Ca-bd_dom
IPR000152 EGF-type_Asp/Asn_hydroxyl_site
IPR017975 Tubulin_CS
IPR000859 CUB_dom
IPR036445 GPCR_2_extracell_dom_sf
IPR013032 EGF-like_CS
IPR000742 EGF-like_dom
IPR036179 Ig-like_dom_sf
IPR001879 GPCR_2_extracellular_dom
IPR003599 Ig_sub
IPR007110 Ig-like_dom
IPR013783 Ig-like_fold
IPR003598 Ig_sub2
IPR000203 GPS
IPR029064 L30e-like
IPR001881 EGF-like_Ca-bd_dom
IPR000152 EGF-type_Asp/Asn_hydroxyl_site
IPR017975 Tubulin_CS
CDD
ProteinModelPortal
H9J5Q1
A0A0L7L2F1
A0A194RQH7
A0A2A4JRC1
A0A2H1X184
A0A194PFQ5
+ More
A0A2W1BIR1 A0A1A9WKE3 A0A182JB14 A0A1B0DAM9 A0A1B0GA86 A0A0L0CG37 A0A182GYS3 Q16H97 Q16ZC4 A0A1B0AJS1 A0A1A9UPM5 A0A1A9XN51 A0A1B0AT93 A0A182RW89 A0A084WG67 A0A1I8PU15 A0A182WGN2 A0A182YDF5 A0A182SZP6 A0A182XGA9 Q7Q1G8 A0A182HRT0 A0A182JY43 B0WGS9 A0A182UFP0 A0A182MN39 A0A182V1X9 A0A1I8M3C5 A0A182KTX1 B4G7L8 A0A182PCU2 A0A154PRY8 A0A2R7WQP9 Q29NG8 A0A182NBV7 A0A3B0JU17 B3N920 A0A1J1I3T9 A0A182QIC5 A0A2A3ELN7 B3MPK9 A0A0P8XGM3 A0A0P8Y4H3 E1ZWL7 A0A088A1Y9 W8BT52 W5JFA1 A0A1W4UFM5 A0A0R1DLB3 T1HCX1 A0A195F423 B4Q8P5 A0A195D9U6 A0A195CEW3 A0A0J7KUZ6 A0A0L7QUB9 E2BQD5 B4HWG3 A0A151XEB2 A0A182F2Y7 A0A2P8Z8P5 A0A195BP00 A0A139WC50 B4JCJ1 F4W506 A0A1B6ILL7 A0A0J9QZQ8 A0A158NVD3 Q9VL42 B4KH51 E9J311 A0A1B6CZL9 A0A0M9AA20 A0A0A9VU98 A0A146LK95 A0A3L8DNQ9 B4NLH4 B4M9D6 A0A2R7WPD6 K7IX39 A0A026X531 T1HCX0 E0V9X9 A0A2P6KAI8 R7VIL3
A0A2W1BIR1 A0A1A9WKE3 A0A182JB14 A0A1B0DAM9 A0A1B0GA86 A0A0L0CG37 A0A182GYS3 Q16H97 Q16ZC4 A0A1B0AJS1 A0A1A9UPM5 A0A1A9XN51 A0A1B0AT93 A0A182RW89 A0A084WG67 A0A1I8PU15 A0A182WGN2 A0A182YDF5 A0A182SZP6 A0A182XGA9 Q7Q1G8 A0A182HRT0 A0A182JY43 B0WGS9 A0A182UFP0 A0A182MN39 A0A182V1X9 A0A1I8M3C5 A0A182KTX1 B4G7L8 A0A182PCU2 A0A154PRY8 A0A2R7WQP9 Q29NG8 A0A182NBV7 A0A3B0JU17 B3N920 A0A1J1I3T9 A0A182QIC5 A0A2A3ELN7 B3MPK9 A0A0P8XGM3 A0A0P8Y4H3 E1ZWL7 A0A088A1Y9 W8BT52 W5JFA1 A0A1W4UFM5 A0A0R1DLB3 T1HCX1 A0A195F423 B4Q8P5 A0A195D9U6 A0A195CEW3 A0A0J7KUZ6 A0A0L7QUB9 E2BQD5 B4HWG3 A0A151XEB2 A0A182F2Y7 A0A2P8Z8P5 A0A195BP00 A0A139WC50 B4JCJ1 F4W506 A0A1B6ILL7 A0A0J9QZQ8 A0A158NVD3 Q9VL42 B4KH51 E9J311 A0A1B6CZL9 A0A0M9AA20 A0A0A9VU98 A0A146LK95 A0A3L8DNQ9 B4NLH4 B4M9D6 A0A2R7WPD6 K7IX39 A0A026X531 T1HCX0 E0V9X9 A0A2P6KAI8 R7VIL3
PDB
6GJE
E-value=0.000413316,
Score=96
Ontologies
Topology
SignalP
Position: 1 - 17,
Likelihood: 0.996788
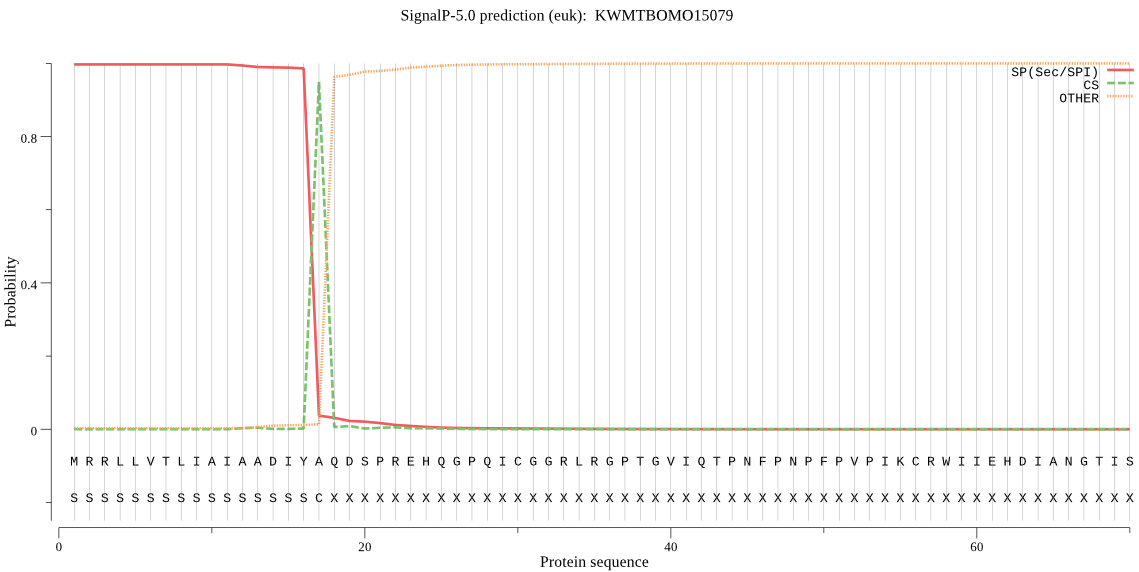
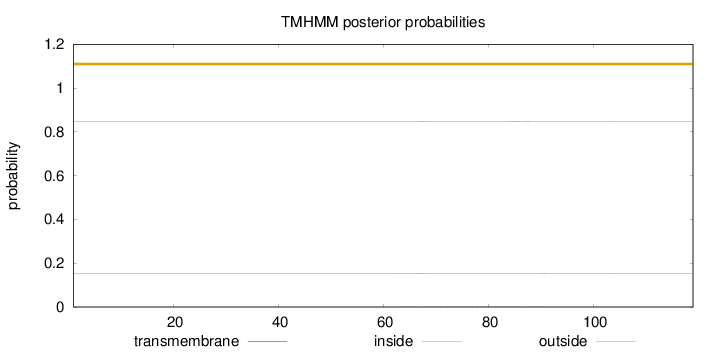
Length:
119
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.03529
Exp number, first 60 AAs:
0.00107
Total prob of N-in:
0.15353
outside
1 - 119
Population Genetic Test Statistics
Pi
75.282224
Theta
93.427933
Tajima's D
-0.630442
CLR
24.796442
CSRT
0.214489275536223
Interpretation
Uncertain
