Gene
KWMTBOMO15076
Pre Gene Modal
BGIBMGA004844
Annotation
PREDICTED:_uncharacterized_protein_LOC101741141_isoform_X3_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.248 PlasmaMembrane Reliability : 1.083
Sequence
CDS
ATGTGGGCCGCCAGACGTGACGCAGGAACCTTGAGGACGGTGCTAGCTAGACTGGCGGCAATGAGGACTCGTACTGACAGGCTCCGCTTGCTCCCCGCTACCCTGCACTTCGATATGCAACCAGCGCTCAACCTACACGCATTAATCGTAAATCAACGTCCGGAAGTCTGGGAGGGTTCAGAGTTCATTCTGAGCTGCATGGCTTACGGATCTCCACATATCGCCTTCACTTGGTACAAAGATGGAGTCAAAATCAATTTTAATGGAACAACAAGAGAAATCTGGACTAGGACAGTCTCAGAAGACGCCCTCGGTCGTCGTATGTCGGTACTCGGAGTAGCTGAAGCCAAAAGACTGGACGGCGGCAAGTGGTCCTGCACCGCTGATGACGCCGGCAGAAGACGTTGCAGAGCCCTAATGCTGGCAGTCCTGAGACCACCGAATATACGGCTTGTACCTTCCACTTTAACAGTAAACAAGGGAGAAAACGTGAGCATCACGTGTTTGGCAGGAGCAAGTAGAATGCACGGTGCTTTGGGGTTCAGTTGGGCTCGAGAACGCGGTCTTTTACCTCTGAATCCAGGTCGAGAAGTGTGGGAAGACTTGTACCCTGCTGGAAGTGTGTTGAAATTGTATAACGTTCAGAAATCTGGTGAATTCCGTTGCCAAGTATCTTCAATAGCCGGTACTAACTCTAAGGGAGTAACAATGTGGGCTCTTGGAACTAAAGACGAGGCTTGTGAAAGTGACGCTTCGCATGGTCTCCGATGGCCGAAAACTGCCCCTGGTGCTCATGCCTCAGCGTCTTGCCCTCCTGGACACACTGGAGAAACTACTAGATTCTGCGAACCGAAAACGACCCAGCACGGTGTTAAATGGAAGCTACCAGATTTCTCCGGATGCGTCGCGGATTCATTAAACGACATTTACGAGCAATTCACTCACATCTACTTTGGGTATTCCTGGAATAACGTATCGAACGTAGCCCGTCAGTATGGAGCGGTCCTTCGCTCCCTACCATCACATCCAGGCGAAGGGACGCTTCCCCTCCACCAGACAAAAGAAATGCTGCACTATTTGTTATCTTCCGCTGGACGACCGGCAGACAGAATGCAAAGTGCTGAGCATCTTCTCAAGATATACGACACTTTGCTCAAGCATCCTGATGCCTTTTTGGATGAAGAGAAAGTCTACGATCTTCAAAACTCGATAGCAGAGACAGCCGGAATGCGAACCAACCTCGACACACATTTCCACGAGTTTACAGTCAAAACGAAACCAGCGAGAGAAGACAGTGCTGCACATTTTGATCTTCAGTCAGGTCCTATTGGATCTGAGGAGTGGATGCTGACGTCAGCCGGCACTGAATTACTCGCTAGAACGGGAAACGCTACCGTTGTTGCAGTATTGTATCACAATTTGGGAACAAGATTACCTTCTTTAAGGCGATCTGTAGAGTATAACCAGTCGTCTTCTAGGAGCGGTCGTGAAATGGAGTATCTGGTGTCATCCCAACAGATCCAATTGTACGCCACTGGTCCCGGAGTCGACAATAGCTTACAACATACGGTGAACCTATTATTCACTCATGTCAAGAACTATAGCGCTGTTGCATCAAAACTCGCCTGTGGTCTTCGCACTCCGTCTGACCCAAGAACATGGATCACCAAAGCTTGTGAAGTGAAAGTTCCTGAAGCAACGCACGTGGCTTGCAAATGTCGAGGGCTTGGTACATATGCACTATTCACGATAGCCAGATCTACGTTGACCGAGGCAGAAAAGGACCTGCGCGGTATTGTGAAGATAACGGTCAGTCTGGGAGGAGCGATGTGCTTAGCTGCAGCAGCTTTGCAACTGCTGGCACTGATTGCTGGGACTAAGGCGAAGCTACCTGTACTACTGAAAGCTGCGACTGCGGGGACTCACTCAGCTGCGATGCTGACCCTGCTAGAGTGTGACACAAGACAGGATGAAGCGTGTCCGGGTACTCTGGGATGGGTGTGCGCGGCGTGCTGGTGCGCGGGGTGCGCGGCGCTTTGTGCGCAGCCACTACTGCTGCAGGCCGAGCTCGCTGGTAGAGCTCAGAAGGCACCGTCTGTCGGACTACTTGCCGGTGTGTGCACATTGTCGTGGCTGACGGCACGGATATGGGGCGGCGCGCCCCTGCAAGTGGGGGCGGGTGCGGCCGCAGTGTGCGCGGCAGGATGCGCGCTACTCGCCTCTCTCTGCTTGGCGCTAGCACTGTGCGCCGCTGCGAGACTGCGAGCCATCTCACACAAGGTCCCCGCTGATAGGAGAAGCTATATCAATGACAGAGGGCGCGTCGTTCGTCACACATTAATACTTCTAGTGTCAACATCGGCAGCTCAAGCAGCGGGTGTGGCTTACGCACAGCCCGGGGAACGGACTGTGGCGCACGTCGCCGCGCTATCCATCGCAGCCCTGGCCAACGGTATAGCGATGCTGGCTTGCTACGTGATAAGAGATGAAGAGTGTCTGCGTTCAGTCCGAAAAGTCCTGACACTGACTCCGGCTCGAGACTGGAGAGATGAACCCACCGGAGATACCTCGCTGAGTTTATACATAAAACAAGGAGGTGAAGTTGAGAGTCGCGGCGGCGTCGGCATATTTGAAAGCACATCGTCACCGGTTTCACCAATTTCCTCATACTGGCGAGCTCCTGGACTTGAAAGCCCTACTAGATTACACAGGCGTCAATCGACTCCAGACAGTCGTGCCGACATCGTCCGTTGTGTAGAATATAAGTGTCCAGTGAGAGGGCCATCGGGTGGACAAGCTGTCTGCGACCTCATCCCTCGCGGCGAGTACGACGCACGTGTGTGTCTAGAGCTCGGTGTGATGCGCCCTCCACCGCCCCCCACCCTCCTCACCTGCTCCCTCGACATCGAACCCAACAAGCGAATCGTCCACCCCACCGACACGTGCACCATTTGCGTCCACTCCAATCCGGACGTCTCTAAAATTCCCTCGCCTCCAATCAAAAGCTGCCTCAAAAAAAATAAGAATCTATCTTCGAGCTCCTCCCTGCCATCCATCGAGGCCAGCAAATCCGATATCGACCAGATCAACAGAGAATGGAAGAAAGCCTACGATTCCCCGGAAACTGATAAAATGCTCAACAAAATCTCGAGCGATCTGGATTTTCTATTGAATAGGAATCAAGAGAGCAAAGCCGCTCTCGACCAAATAGAAGAGGCTCCGACTTAG
Protein
MWAARRDAGTLRTVLARLAAMRTRTDRLRLLPATLHFDMQPALNLHALIVNQRPEVWEGSEFILSCMAYGSPHIAFTWYKDGVKINFNGTTREIWTRTVSEDALGRRMSVLGVAEAKRLDGGKWSCTADDAGRRRCRALMLAVLRPPNIRLVPSTLTVNKGENVSITCLAGASRMHGALGFSWARERGLLPLNPGREVWEDLYPAGSVLKLYNVQKSGEFRCQVSSIAGTNSKGVTMWALGTKDEACESDASHGLRWPKTAPGAHASASCPPGHTGETTRFCEPKTTQHGVKWKLPDFSGCVADSLNDIYEQFTHIYFGYSWNNVSNVARQYGAVLRSLPSHPGEGTLPLHQTKEMLHYLLSSAGRPADRMQSAEHLLKIYDTLLKHPDAFLDEEKVYDLQNSIAETAGMRTNLDTHFHEFTVKTKPAREDSAAHFDLQSGPIGSEEWMLTSAGTELLARTGNATVVAVLYHNLGTRLPSLRRSVEYNQSSSRSGREMEYLVSSQQIQLYATGPGVDNSLQHTVNLLFTHVKNYSAVASKLACGLRTPSDPRTWITKACEVKVPEATHVACKCRGLGTYALFTIARSTLTEAEKDLRGIVKITVSLGGAMCLAAAALQLLALIAGTKAKLPVLLKAATAGTHSAAMLTLLECDTRQDEACPGTLGWVCAACWCAGCAALCAQPLLLQAELAGRAQKAPSVGLLAGVCTLSWLTARIWGGAPLQVGAGAAAVCAAGCALLASLCLALALCAAARLRAISHKVPADRRSYINDRGRVVRHTLILLVSTSAAQAAGVAYAQPGERTVAHVAALSIAALANGIAMLACYVIRDEECLRSVRKVLTLTPARDWRDEPTGDTSLSLYIKQGGEVESRGGVGIFESTSSPVSPISSYWRAPGLESPTRLHRRQSTPDSRADIVRCVEYKCPVRGPSGGQAVCDLIPRGEYDARVCLELGVMRPPPPPTLLTCSLDIEPNKRIVHPTDTCTICVHSNPDVSKIPSPPIKSCLKKNKNLSSSSSLPSIEASKSDIDQINREWKKAYDSPETDKMLNKISSDLDFLLNRNQESKAALDQIEEAPT
Summary
Pubmed
EMBL
Proteomes
Interpro
Gene 3D
ProteinModelPortal
PDB
4YFG
E-value=4.80657e-06,
Score=124
Ontologies
GO
Topology
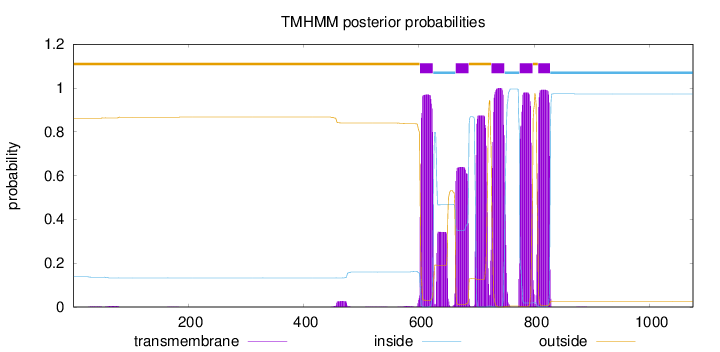
Length:
1075
Number of predicted TMHs:
5
Exp number of AAs in TMHs:
128.30058
Exp number, first 60 AAs:
0.03919
Total prob of N-in:
0.13776
outside
1 - 601
TMhelix
602 - 624
inside
625 - 663
TMhelix
664 - 686
outside
687 - 725
TMhelix
726 - 748
inside
749 - 774
TMhelix
775 - 797
outside
798 - 806
TMhelix
807 - 827
inside
828 - 1075
Population Genetic Test Statistics
Pi
260.998195
Theta
196.246634
Tajima's D
0.781202
CLR
0.172794
CSRT
0.59822008899555
Interpretation
Uncertain
