Pre Gene Modal
BGIBMGA004828
Annotation
PREDICTED:_lysM_and_putative_peptidoglycan-binding_domain-containing_protein_2_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 3.939
Sequence
CDS
ATGTCATATTTAAAGACGTCAAATATGGAGGAGGAACGTCTCTTTGACGAGAGAGTTTCTATTCGAGAGTCTGCTCGACCATTAAAAAAGTATGGAAGCACCTGCAACCACATGAAAAGGAACGAGGAATACATTAAACATGTTATGTCTAAAGGGGACACCTTGCAAGGGATCGCATTAAAATATGGTGTAACTATGGAGAAAATAAGAAGGGCCAATAGATTGTTCGCTACGGACAGTTTGTTTTTGAGAGAATATTTATTGATTCCGGTGACCAAAGATTCTCCTTTTTACGAAAACGGGGAACGCGTAACCGAGCTGCCTCCTCCGAATCATCGCGCCTCCATAGCAGGCATGCCAACTAGAAACAGTTTTTCGCCTGGAAGCCCCGATGAGAAGAAGAGTTTTGACGAATTCCTGAACAGAGTAGACTCATCATTGGCTGACATAAAGCGACATGTAGAAAAAACAAAAGAAAACAGTGAGTTCGTGAAGGATATAGATGAGGGTTACGTGAGGCACAGGCCGACGGCGCGACTCAGACAGTCCGCGTCTATGGGCGCTTCGTCGTCTTACAATGAAGTAATACCGCCTCCCTTGCCGCCTCCGCCCGTCTCCCTCACCCAGGGCAGACGCGTCGACTCCTCGCTCAGACGGCTGGAGAGGACACACGACGACCTATTCCAGTTGTAG
Protein
MSYLKTSNMEEERLFDERVSIRESARPLKKYGSTCNHMKRNEEYIKHVMSKGDTLQGIALKYGVTMEKIRRANRLFATDSLFLREYLLIPVTKDSPFYENGERVTELPPPNHRASIAGMPTRNSFSPGSPDEKKSFDEFLNRVDSSLADIKRHVEKTKENSEFVKDIDEGYVRHRPTARLRQSASMGASSSYNEVIPPPLPPPPVSLTQGRRVDSSLRRLERTHDDLFQL
Summary
Uniprot
A0A2W1BW63
A0A2A4JGP8
A0A3S2M9D7
A0A212FLH8
A0A194PM71
A0A2P8ZF91
+ More
A0A0K8TMA5 A0A2J7R2U3 A0A195B408 A0A1B0D8H2 A0A0L7RI84 E2BKP7 A0A158NTD8 A0A1B6HND2 A0A151IY23 A0A2A3E1U5 F4W3Y9 A0A088AAE6 A0A151JTL5 A0A232ERJ8 A0A0N0U6W8 A0A151X1E4 A0A0T6B560 A0A195CJX2 A0A026VVB0 A0A154PCP3 A0A1L8E3R5 E9I9R0 U5ERF8 E2APP0 R4WPT8 A0A1Y1LL00 D6WU87 A0A0A9WVW5 A0A1B6C969 A0A3L8DQL3 A0A1J1HNZ0 A0A1L8E3R7 A0A1L8E3Y9 N6TQ23 B5DSS7 A0A1S4F0W6 Q17J12 A0A0J7KL67 E0VUG1 A0A224XUL6 A0A0V0GA36 A0A023F7Z6 R4G5D2 A0A1Q3FVG9 B0WJG1 A0A182QB98 A0A182VYU0 A0A2M4ASC7 A0A2S2QFQ4 W5JIV5
A0A0K8TMA5 A0A2J7R2U3 A0A195B408 A0A1B0D8H2 A0A0L7RI84 E2BKP7 A0A158NTD8 A0A1B6HND2 A0A151IY23 A0A2A3E1U5 F4W3Y9 A0A088AAE6 A0A151JTL5 A0A232ERJ8 A0A0N0U6W8 A0A151X1E4 A0A0T6B560 A0A195CJX2 A0A026VVB0 A0A154PCP3 A0A1L8E3R5 E9I9R0 U5ERF8 E2APP0 R4WPT8 A0A1Y1LL00 D6WU87 A0A0A9WVW5 A0A1B6C969 A0A3L8DQL3 A0A1J1HNZ0 A0A1L8E3R7 A0A1L8E3Y9 N6TQ23 B5DSS7 A0A1S4F0W6 Q17J12 A0A0J7KL67 E0VUG1 A0A224XUL6 A0A0V0GA36 A0A023F7Z6 R4G5D2 A0A1Q3FVG9 B0WJG1 A0A182QB98 A0A182VYU0 A0A2M4ASC7 A0A2S2QFQ4 W5JIV5
Pubmed
EMBL
KZ149918
PZC77854.1
NWSH01001594
PCG70764.1
RSAL01000008
RVE53954.1
+ More
AGBW02007763 OWR54591.1 KQ459604 KPI92210.1 PYGN01000075 PSN55169.1 GDAI01002325 JAI15278.1 NEVH01007825 PNF35159.1 KQ976625 KYM78944.1 AJVK01012625 AJVK01012626 KQ414585 KOC70539.1 GL448819 EFN83745.1 ADTU01025848 GECU01031509 JAS76197.1 KQ980778 KYN13212.1 KZ288444 PBC25665.1 GL887491 EGI70985.1 KQ981885 KYN33865.1 NNAY01002610 OXU20952.1 KQ435727 KOX77982.1 KQ982585 KYQ54222.1 LJIG01009775 KRT82406.1 KQ977642 KYN01026.1 KK107847 EZA47471.1 KQ434870 KZC09557.1 GFDF01000707 JAV13377.1 GL761846 EFZ22647.1 GANO01003767 JAB56104.1 GL441572 EFN64591.1 AK417686 BAN20901.1 GEZM01054515 JAV73591.1 KQ971352 EFA06782.1 GBHO01031690 GBHO01016359 GBRD01006260 JAG11914.1 JAG27245.1 JAG59561.1 GEDC01027297 JAS10001.1 QOIP01000005 RLU22552.1 CVRI01000011 CRK89250.1 GFDF01000706 JAV13378.1 GFDF01000708 JAV13376.1 APGK01025694 KB740600 KB631982 ENN80108.1 ERL87601.1 CH475602 EDY70846.2 CH477235 EAT46699.1 LBMM01006052 KMQ90981.1 DS235786 EEB17017.1 GFTR01004234 JAW12192.1 GECL01001407 JAP04717.1 GBBI01001349 JAC17363.1 ACPB03015538 GAHY01000645 JAA76865.1 GFDL01003435 JAV31610.1 DS231959 EDS29135.1 AXCN02001171 GGFK01010362 MBW43683.1 GGMS01007351 MBY76554.1 ADMH02001372 ETN62724.1
AGBW02007763 OWR54591.1 KQ459604 KPI92210.1 PYGN01000075 PSN55169.1 GDAI01002325 JAI15278.1 NEVH01007825 PNF35159.1 KQ976625 KYM78944.1 AJVK01012625 AJVK01012626 KQ414585 KOC70539.1 GL448819 EFN83745.1 ADTU01025848 GECU01031509 JAS76197.1 KQ980778 KYN13212.1 KZ288444 PBC25665.1 GL887491 EGI70985.1 KQ981885 KYN33865.1 NNAY01002610 OXU20952.1 KQ435727 KOX77982.1 KQ982585 KYQ54222.1 LJIG01009775 KRT82406.1 KQ977642 KYN01026.1 KK107847 EZA47471.1 KQ434870 KZC09557.1 GFDF01000707 JAV13377.1 GL761846 EFZ22647.1 GANO01003767 JAB56104.1 GL441572 EFN64591.1 AK417686 BAN20901.1 GEZM01054515 JAV73591.1 KQ971352 EFA06782.1 GBHO01031690 GBHO01016359 GBRD01006260 JAG11914.1 JAG27245.1 JAG59561.1 GEDC01027297 JAS10001.1 QOIP01000005 RLU22552.1 CVRI01000011 CRK89250.1 GFDF01000706 JAV13378.1 GFDF01000708 JAV13376.1 APGK01025694 KB740600 KB631982 ENN80108.1 ERL87601.1 CH475602 EDY70846.2 CH477235 EAT46699.1 LBMM01006052 KMQ90981.1 DS235786 EEB17017.1 GFTR01004234 JAW12192.1 GECL01001407 JAP04717.1 GBBI01001349 JAC17363.1 ACPB03015538 GAHY01000645 JAA76865.1 GFDL01003435 JAV31610.1 DS231959 EDS29135.1 AXCN02001171 GGFK01010362 MBW43683.1 GGMS01007351 MBY76554.1 ADMH02001372 ETN62724.1
Proteomes
UP000218220
UP000283053
UP000007151
UP000053268
UP000245037
UP000235965
+ More
UP000078540 UP000092462 UP000053825 UP000008237 UP000005205 UP000078492 UP000242457 UP000007755 UP000005203 UP000078541 UP000215335 UP000053105 UP000075809 UP000078542 UP000053097 UP000076502 UP000000311 UP000007266 UP000279307 UP000183832 UP000019118 UP000030742 UP000001819 UP000008820 UP000036403 UP000009046 UP000015103 UP000002320 UP000075886 UP000075920 UP000000673
UP000078540 UP000092462 UP000053825 UP000008237 UP000005205 UP000078492 UP000242457 UP000007755 UP000005203 UP000078541 UP000215335 UP000053105 UP000075809 UP000078542 UP000053097 UP000076502 UP000000311 UP000007266 UP000279307 UP000183832 UP000019118 UP000030742 UP000001819 UP000008820 UP000036403 UP000009046 UP000015103 UP000002320 UP000075886 UP000075920 UP000000673
Pfam
PF01476 LysM
Gene 3D
CDD
ProteinModelPortal
A0A2W1BW63
A0A2A4JGP8
A0A3S2M9D7
A0A212FLH8
A0A194PM71
A0A2P8ZF91
+ More
A0A0K8TMA5 A0A2J7R2U3 A0A195B408 A0A1B0D8H2 A0A0L7RI84 E2BKP7 A0A158NTD8 A0A1B6HND2 A0A151IY23 A0A2A3E1U5 F4W3Y9 A0A088AAE6 A0A151JTL5 A0A232ERJ8 A0A0N0U6W8 A0A151X1E4 A0A0T6B560 A0A195CJX2 A0A026VVB0 A0A154PCP3 A0A1L8E3R5 E9I9R0 U5ERF8 E2APP0 R4WPT8 A0A1Y1LL00 D6WU87 A0A0A9WVW5 A0A1B6C969 A0A3L8DQL3 A0A1J1HNZ0 A0A1L8E3R7 A0A1L8E3Y9 N6TQ23 B5DSS7 A0A1S4F0W6 Q17J12 A0A0J7KL67 E0VUG1 A0A224XUL6 A0A0V0GA36 A0A023F7Z6 R4G5D2 A0A1Q3FVG9 B0WJG1 A0A182QB98 A0A182VYU0 A0A2M4ASC7 A0A2S2QFQ4 W5JIV5
A0A0K8TMA5 A0A2J7R2U3 A0A195B408 A0A1B0D8H2 A0A0L7RI84 E2BKP7 A0A158NTD8 A0A1B6HND2 A0A151IY23 A0A2A3E1U5 F4W3Y9 A0A088AAE6 A0A151JTL5 A0A232ERJ8 A0A0N0U6W8 A0A151X1E4 A0A0T6B560 A0A195CJX2 A0A026VVB0 A0A154PCP3 A0A1L8E3R5 E9I9R0 U5ERF8 E2APP0 R4WPT8 A0A1Y1LL00 D6WU87 A0A0A9WVW5 A0A1B6C969 A0A3L8DQL3 A0A1J1HNZ0 A0A1L8E3R7 A0A1L8E3Y9 N6TQ23 B5DSS7 A0A1S4F0W6 Q17J12 A0A0J7KL67 E0VUG1 A0A224XUL6 A0A0V0GA36 A0A023F7Z6 R4G5D2 A0A1Q3FVG9 B0WJG1 A0A182QB98 A0A182VYU0 A0A2M4ASC7 A0A2S2QFQ4 W5JIV5
PDB
2DJP
E-value=3.38837e-10,
Score=153
Ontologies
GO
Topology
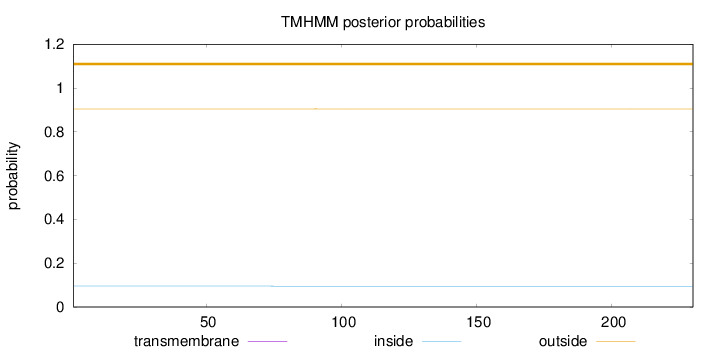
Length:
230
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00328
Exp number, first 60 AAs:
0.00105
Total prob of N-in:
0.09546
outside
1 - 230
Population Genetic Test Statistics
Pi
325.583277
Theta
183.826113
Tajima's D
2.713585
CLR
0.229538
CSRT
0.963101844907755
Interpretation
Uncertain
