Pre Gene Modal
BGIBMGA004846
Annotation
small_GTP_binding_protein_RAB8_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.243 Mitochondrial Reliability : 1.166
Sequence
CDS
ATGGCAAAAACTTACGATTGTTTATTCAAATTACTTCTAATCGGAGATTCGGGAGTGGGAAAAACATCAATACTATTTAGATTTTCTGAAGATGCATTTAACATCTCCTTTATATCCACAATTGGTATCGACTTTAAAATTCGAACAATAGACCTTGATGGCAAAAAAGTTAAATTACAAATATGGGATACTGCTGGTCAAGAGAGATTCCGTACAATAACAACAGCATACTACCGTGGTTCTATGGGAATTATGCTTGTCTACGATGTTACGAATGAGAAGAGCTTTGAAAACATTAAGAATTGGATTAGGAACATTGAAGAGAATGCCAGTGCTGATGTTGAGAAAATGATACTTGGAAATAAGTGTGACTTAGATTCTCAAAGACAGGTATCAAAGGAGCGAGGTGAACAATTAGCCATAGAATATCAGATAAAATTTGTGGAGACATCAGCAAAAGACTCGTTAAATGTTGAATATGCATTTTACACTTTAGCCAGGGACATAAAAGCGAAAATGGAAAAGAAACAGGAAGCGAGTAATCCGTCGGGCGGTAGGAGTGGCGTCCACAAGCCGACGCTGAACGACGCTCGCAAGCCGGTGACGTGGCTGTCGCGTTGCAGCGTGCTTTGA
Protein
MAKTYDCLFKLLLIGDSGVGKTSILFRFSEDAFNISFISTIGIDFKIRTIDLDGKKVKLQIWDTAGQERFRTITTAYYRGSMGIMLVYDVTNEKSFENIKNWIRNIEENASADVEKMILGNKCDLDSQRQVSKERGEQLAIEYQIKFVETSAKDSLNVEYAFYTLARDIKAKMEKKQEASNPSGGRSGVHKPTLNDARKPVTWLSRCSVL
Summary
Uniprot
Q1HQB6
A0A0N1I9D0
A0A194PM64
A0A212FLH4
S4PY78
A0A2A4JDF2
+ More
A0A2H1V4E6 I4DNS8 H9J5Q6 A0A0M8ZQ36 A0A0L7R0D1 G3LF44 A0A154NZQ5 A0A0J7LAH2 D6WP52 A0A195EFP1 F4WGA8 A0A195B6Y5 A0A151JY29 E2AZF2 A0A026WTL0 A0A151WJW1 N6SZJ3 V5IAH0 E2B8I5 A0A3L8D9Q1 A0A195CPJ3 A0A232FIJ0 K7J1J1 A0A2J7PV62 E9GUJ7 A0A0P5R8P7 A0A1A9XDC5 A0A1B0C0B7 A0A1A9X110 D3TQA6 A0A1B6DXA7 A0A1I8M0L0 A0A1I8QEZ2 A0A1L8EH47 A0A2W1BU31 A0A0L0CFD5 A0A0C9QCZ7 A0A1Y1NF54 A0A165VFU0 A0A1B6GTU6 A0A1B6LJF8 A0A1B0C8M2 A0A1L8DV34 A0A1L8DV33 W8BD57 A0A0K8VWG5 A0A034VUG5 A0A0K8TS87 B4MLG7 A0A084W017 A0A182JN04 A0A0A1WWU9 A0A0P4WAV4 U5EZ96 B4LBM1 B4L0W9 A0A0M4EM34 A0A3B0JU77 B4H930 B5DPR3 B4IXP2 B3M8T8 B4IIR8 B3NDZ9 B4QRC2 O18338 B4PFW9 A0A182NQT1 J9JN61 A0A0L7L849 A0A1W4VU41 A0A023F5S8 A0A0P4W1W9 T1E2D6 A0A2S2Q7E1 A0A0A9VV46 A0A182YHW3 T1DP62 A0A2M3ZHX1 A0A2M4AN18 W5J8J8 A0A182FS40 A0A069DXB4 A0A023ELP6 A0A1Q3FWZ4 Q16TQ1 A0A2H8TZG0 A0A182X9B8 A0A3F2YUQ3 A0A182PRW4 A0A182Q2V0 A0A182UXJ3 A0A182K6B0 Q7PUB2
A0A2H1V4E6 I4DNS8 H9J5Q6 A0A0M8ZQ36 A0A0L7R0D1 G3LF44 A0A154NZQ5 A0A0J7LAH2 D6WP52 A0A195EFP1 F4WGA8 A0A195B6Y5 A0A151JY29 E2AZF2 A0A026WTL0 A0A151WJW1 N6SZJ3 V5IAH0 E2B8I5 A0A3L8D9Q1 A0A195CPJ3 A0A232FIJ0 K7J1J1 A0A2J7PV62 E9GUJ7 A0A0P5R8P7 A0A1A9XDC5 A0A1B0C0B7 A0A1A9X110 D3TQA6 A0A1B6DXA7 A0A1I8M0L0 A0A1I8QEZ2 A0A1L8EH47 A0A2W1BU31 A0A0L0CFD5 A0A0C9QCZ7 A0A1Y1NF54 A0A165VFU0 A0A1B6GTU6 A0A1B6LJF8 A0A1B0C8M2 A0A1L8DV34 A0A1L8DV33 W8BD57 A0A0K8VWG5 A0A034VUG5 A0A0K8TS87 B4MLG7 A0A084W017 A0A182JN04 A0A0A1WWU9 A0A0P4WAV4 U5EZ96 B4LBM1 B4L0W9 A0A0M4EM34 A0A3B0JU77 B4H930 B5DPR3 B4IXP2 B3M8T8 B4IIR8 B3NDZ9 B4QRC2 O18338 B4PFW9 A0A182NQT1 J9JN61 A0A0L7L849 A0A1W4VU41 A0A023F5S8 A0A0P4W1W9 T1E2D6 A0A2S2Q7E1 A0A0A9VV46 A0A182YHW3 T1DP62 A0A2M3ZHX1 A0A2M4AN18 W5J8J8 A0A182FS40 A0A069DXB4 A0A023ELP6 A0A1Q3FWZ4 Q16TQ1 A0A2H8TZG0 A0A182X9B8 A0A3F2YUQ3 A0A182PRW4 A0A182Q2V0 A0A182UXJ3 A0A182K6B0 Q7PUB2
Pubmed
26354079
22118469
23622113
22651552
19121390
18362917
+ More
19820115 21719571 20798317 24508170 23537049 30249741 28648823 20075255 21292972 20353571 25315136 28756777 26108605 28004739 24495485 25348373 26369729 17994087 24438588 25830018 15632085 23185243 22936249 9074639 10731132 12537568 12537572 12537573 12537574 14722103 16110336 17569856 17569867 26109357 26109356 17550304 26227816 25474469 27129103 24330624 25401762 25244985 20920257 23761445 26334808 24945155 17510324 20966253 12364791 14747013 17210077
19820115 21719571 20798317 24508170 23537049 30249741 28648823 20075255 21292972 20353571 25315136 28756777 26108605 28004739 24495485 25348373 26369729 17994087 24438588 25830018 15632085 23185243 22936249 9074639 10731132 12537568 12537572 12537573 12537574 14722103 16110336 17569856 17569867 26109357 26109356 17550304 26227816 25474469 27129103 24330624 25401762 25244985 20920257 23761445 26334808 24945155 17510324 20966253 12364791 14747013 17210077
EMBL
DQ443136
DQ525203
ABF51225.1
ABF71468.1
KQ460594
KPJ13917.1
+ More
KQ459604 KPI92205.1 AGBW02007763 OWR54594.1 GAIX01003763 JAA88797.1 NWSH01001797 PCG70117.1 ODYU01000443 SOQ35222.1 AK403119 BAM19568.1 BABH01025026 KQ436240 KOX67343.1 KQ414670 KOC64302.1 JN315689 AEO51738.1 KQ434781 KZC04478.1 LBMM01000112 KMR04887.1 KQ971352 EFA07283.1 KQ978957 KYN27063.1 GL888128 EGI66875.1 KQ976579 KYM80022.1 KQ981578 KYN39756.1 GL444207 EFN61191.1 KK107111 EZA59001.1 KQ983039 KYQ47985.1 APGK01058332 KB741288 ENN70678.1 GALX01001200 JAB67266.1 GL446334 EFN87994.1 QOIP01000011 RLU17235.1 KQ977481 KYN02412.1 NNAY01000155 OXU30492.1 NEVH01020984 PNF20226.1 GL732566 EFX76917.1 GDIQ01111054 GDIP01077266 LRGB01000115 JAL40672.1 JAM26449.1 KZS20766.1 JXJN01023506 CCAG010002318 EZ423608 ADD19884.1 GEDC01006986 JAS30312.1 GFDG01000853 JAV17946.1 KZ149918 PZC77851.1 JRES01000484 KNC30902.1 GBYB01001189 JAG70956.1 GEZM01003834 JAV96532.1 KT984795 AMZ00347.1 GECZ01003923 JAS65846.1 GEBQ01016238 GEBQ01011288 JAT23739.1 JAT28689.1 AJWK01001113 GFDF01003793 JAV10291.1 GFDF01003792 JAV10292.1 GAMC01009949 GAMC01009943 JAB96612.1 GDHF01009098 JAI43216.1 GAKP01011971 JAC46981.1 GDAI01000384 JAI17219.1 CH963847 EDW72823.1 ATLV01019081 KE525262 KFB43561.1 AXCP01002644 GBXI01011287 JAD03005.1 GDRN01057067 JAI65745.1 GANO01001189 JAB58682.1 CH940647 EDW70831.1 CH933809 EDW19219.1 CP012525 ALC44635.1 OUUW01000002 SPP77259.1 CH479226 EDW35245.1 CH379069 EDY73603.1 KRT07915.1 KRT07916.1 KRT07917.1 CH916366 EDV97504.1 CH902618 EDV41089.1 CH480844 EDW49839.1 CH954178 EDV52423.1 CM000363 CM002912 EDX11135.1 KMZ00629.1 AE014296 AY069671 D84347 AB112933 AAF49101.1 AAL39816.1 AFH04508.1 BAA21711.1 BAD07038.1 CM000159 EDW95266.1 KRK02338.1 ABLF02030773 JTDY01002387 KOB71519.1 GBBI01002184 JAC16528.1 GDKW01000966 JAI55629.1 GALA01000819 JAA94033.1 GGMS01004470 MBY73673.1 GBHO01044035 GBRD01017495 JAF99568.1 JAG48332.1 GAMD01002215 JAA99375.1 GGFM01007360 MBW28111.1 GGFK01008677 MBW41998.1 ADMH02002101 GGFL01004304 ETN59119.1 MBW68482.1 GBGD01002905 JAC85984.1 GAPW01004409 JAC09189.1 GFDL01002915 JAV32130.1 CH477643 EAT37877.1 EAT37879.1 EJY57852.1 GFXV01007346 MBW19151.1 AXCN02001500 AAAB01008987 EAA01802.5
KQ459604 KPI92205.1 AGBW02007763 OWR54594.1 GAIX01003763 JAA88797.1 NWSH01001797 PCG70117.1 ODYU01000443 SOQ35222.1 AK403119 BAM19568.1 BABH01025026 KQ436240 KOX67343.1 KQ414670 KOC64302.1 JN315689 AEO51738.1 KQ434781 KZC04478.1 LBMM01000112 KMR04887.1 KQ971352 EFA07283.1 KQ978957 KYN27063.1 GL888128 EGI66875.1 KQ976579 KYM80022.1 KQ981578 KYN39756.1 GL444207 EFN61191.1 KK107111 EZA59001.1 KQ983039 KYQ47985.1 APGK01058332 KB741288 ENN70678.1 GALX01001200 JAB67266.1 GL446334 EFN87994.1 QOIP01000011 RLU17235.1 KQ977481 KYN02412.1 NNAY01000155 OXU30492.1 NEVH01020984 PNF20226.1 GL732566 EFX76917.1 GDIQ01111054 GDIP01077266 LRGB01000115 JAL40672.1 JAM26449.1 KZS20766.1 JXJN01023506 CCAG010002318 EZ423608 ADD19884.1 GEDC01006986 JAS30312.1 GFDG01000853 JAV17946.1 KZ149918 PZC77851.1 JRES01000484 KNC30902.1 GBYB01001189 JAG70956.1 GEZM01003834 JAV96532.1 KT984795 AMZ00347.1 GECZ01003923 JAS65846.1 GEBQ01016238 GEBQ01011288 JAT23739.1 JAT28689.1 AJWK01001113 GFDF01003793 JAV10291.1 GFDF01003792 JAV10292.1 GAMC01009949 GAMC01009943 JAB96612.1 GDHF01009098 JAI43216.1 GAKP01011971 JAC46981.1 GDAI01000384 JAI17219.1 CH963847 EDW72823.1 ATLV01019081 KE525262 KFB43561.1 AXCP01002644 GBXI01011287 JAD03005.1 GDRN01057067 JAI65745.1 GANO01001189 JAB58682.1 CH940647 EDW70831.1 CH933809 EDW19219.1 CP012525 ALC44635.1 OUUW01000002 SPP77259.1 CH479226 EDW35245.1 CH379069 EDY73603.1 KRT07915.1 KRT07916.1 KRT07917.1 CH916366 EDV97504.1 CH902618 EDV41089.1 CH480844 EDW49839.1 CH954178 EDV52423.1 CM000363 CM002912 EDX11135.1 KMZ00629.1 AE014296 AY069671 D84347 AB112933 AAF49101.1 AAL39816.1 AFH04508.1 BAA21711.1 BAD07038.1 CM000159 EDW95266.1 KRK02338.1 ABLF02030773 JTDY01002387 KOB71519.1 GBBI01002184 JAC16528.1 GDKW01000966 JAI55629.1 GALA01000819 JAA94033.1 GGMS01004470 MBY73673.1 GBHO01044035 GBRD01017495 JAF99568.1 JAG48332.1 GAMD01002215 JAA99375.1 GGFM01007360 MBW28111.1 GGFK01008677 MBW41998.1 ADMH02002101 GGFL01004304 ETN59119.1 MBW68482.1 GBGD01002905 JAC85984.1 GAPW01004409 JAC09189.1 GFDL01002915 JAV32130.1 CH477643 EAT37877.1 EAT37879.1 EJY57852.1 GFXV01007346 MBW19151.1 AXCN02001500 AAAB01008987 EAA01802.5
Proteomes
UP000053240
UP000053268
UP000007151
UP000218220
UP000005204
UP000053105
+ More
UP000053825 UP000076502 UP000036403 UP000007266 UP000078492 UP000007755 UP000078540 UP000078541 UP000000311 UP000053097 UP000075809 UP000019118 UP000008237 UP000279307 UP000078542 UP000215335 UP000002358 UP000235965 UP000000305 UP000076858 UP000092443 UP000092460 UP000091820 UP000092444 UP000095301 UP000095300 UP000037069 UP000092461 UP000007798 UP000030765 UP000075880 UP000008792 UP000009192 UP000092553 UP000268350 UP000008744 UP000001819 UP000001070 UP000007801 UP000001292 UP000008711 UP000000304 UP000000803 UP000002282 UP000075884 UP000007819 UP000037510 UP000192221 UP000076408 UP000000673 UP000069272 UP000008820 UP000076407 UP000075882 UP000075885 UP000075886 UP000075903 UP000075881 UP000007062
UP000053825 UP000076502 UP000036403 UP000007266 UP000078492 UP000007755 UP000078540 UP000078541 UP000000311 UP000053097 UP000075809 UP000019118 UP000008237 UP000279307 UP000078542 UP000215335 UP000002358 UP000235965 UP000000305 UP000076858 UP000092443 UP000092460 UP000091820 UP000092444 UP000095301 UP000095300 UP000037069 UP000092461 UP000007798 UP000030765 UP000075880 UP000008792 UP000009192 UP000092553 UP000268350 UP000008744 UP000001819 UP000001070 UP000007801 UP000001292 UP000008711 UP000000304 UP000000803 UP000002282 UP000075884 UP000007819 UP000037510 UP000192221 UP000076408 UP000000673 UP000069272 UP000008820 UP000076407 UP000075882 UP000075885 UP000075886 UP000075903 UP000075881 UP000007062
Pfam
PF00071 Ras
Interpro
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
Q1HQB6
A0A0N1I9D0
A0A194PM64
A0A212FLH4
S4PY78
A0A2A4JDF2
+ More
A0A2H1V4E6 I4DNS8 H9J5Q6 A0A0M8ZQ36 A0A0L7R0D1 G3LF44 A0A154NZQ5 A0A0J7LAH2 D6WP52 A0A195EFP1 F4WGA8 A0A195B6Y5 A0A151JY29 E2AZF2 A0A026WTL0 A0A151WJW1 N6SZJ3 V5IAH0 E2B8I5 A0A3L8D9Q1 A0A195CPJ3 A0A232FIJ0 K7J1J1 A0A2J7PV62 E9GUJ7 A0A0P5R8P7 A0A1A9XDC5 A0A1B0C0B7 A0A1A9X110 D3TQA6 A0A1B6DXA7 A0A1I8M0L0 A0A1I8QEZ2 A0A1L8EH47 A0A2W1BU31 A0A0L0CFD5 A0A0C9QCZ7 A0A1Y1NF54 A0A165VFU0 A0A1B6GTU6 A0A1B6LJF8 A0A1B0C8M2 A0A1L8DV34 A0A1L8DV33 W8BD57 A0A0K8VWG5 A0A034VUG5 A0A0K8TS87 B4MLG7 A0A084W017 A0A182JN04 A0A0A1WWU9 A0A0P4WAV4 U5EZ96 B4LBM1 B4L0W9 A0A0M4EM34 A0A3B0JU77 B4H930 B5DPR3 B4IXP2 B3M8T8 B4IIR8 B3NDZ9 B4QRC2 O18338 B4PFW9 A0A182NQT1 J9JN61 A0A0L7L849 A0A1W4VU41 A0A023F5S8 A0A0P4W1W9 T1E2D6 A0A2S2Q7E1 A0A0A9VV46 A0A182YHW3 T1DP62 A0A2M3ZHX1 A0A2M4AN18 W5J8J8 A0A182FS40 A0A069DXB4 A0A023ELP6 A0A1Q3FWZ4 Q16TQ1 A0A2H8TZG0 A0A182X9B8 A0A3F2YUQ3 A0A182PRW4 A0A182Q2V0 A0A182UXJ3 A0A182K6B0 Q7PUB2
A0A2H1V4E6 I4DNS8 H9J5Q6 A0A0M8ZQ36 A0A0L7R0D1 G3LF44 A0A154NZQ5 A0A0J7LAH2 D6WP52 A0A195EFP1 F4WGA8 A0A195B6Y5 A0A151JY29 E2AZF2 A0A026WTL0 A0A151WJW1 N6SZJ3 V5IAH0 E2B8I5 A0A3L8D9Q1 A0A195CPJ3 A0A232FIJ0 K7J1J1 A0A2J7PV62 E9GUJ7 A0A0P5R8P7 A0A1A9XDC5 A0A1B0C0B7 A0A1A9X110 D3TQA6 A0A1B6DXA7 A0A1I8M0L0 A0A1I8QEZ2 A0A1L8EH47 A0A2W1BU31 A0A0L0CFD5 A0A0C9QCZ7 A0A1Y1NF54 A0A165VFU0 A0A1B6GTU6 A0A1B6LJF8 A0A1B0C8M2 A0A1L8DV34 A0A1L8DV33 W8BD57 A0A0K8VWG5 A0A034VUG5 A0A0K8TS87 B4MLG7 A0A084W017 A0A182JN04 A0A0A1WWU9 A0A0P4WAV4 U5EZ96 B4LBM1 B4L0W9 A0A0M4EM34 A0A3B0JU77 B4H930 B5DPR3 B4IXP2 B3M8T8 B4IIR8 B3NDZ9 B4QRC2 O18338 B4PFW9 A0A182NQT1 J9JN61 A0A0L7L849 A0A1W4VU41 A0A023F5S8 A0A0P4W1W9 T1E2D6 A0A2S2Q7E1 A0A0A9VV46 A0A182YHW3 T1DP62 A0A2M3ZHX1 A0A2M4AN18 W5J8J8 A0A182FS40 A0A069DXB4 A0A023ELP6 A0A1Q3FWZ4 Q16TQ1 A0A2H8TZG0 A0A182X9B8 A0A3F2YUQ3 A0A182PRW4 A0A182Q2V0 A0A182UXJ3 A0A182K6B0 Q7PUB2
PDB
5SZI
E-value=1.94521e-84,
Score=793
Ontologies
GO
GO:0003924
GO:0005525
GO:0006886
GO:0017157
GO:0030140
GO:0006904
GO:0008021
GO:0048210
GO:0072659
GO:0005886
GO:0032482
GO:0005768
GO:0032869
GO:0009306
GO:0055037
GO:0045202
GO:0008582
GO:0072657
GO:0043025
GO:0005794
GO:0061865
GO:0016192
GO:0005737
GO:0031982
GO:0007264
GO:0005634
GO:0006260
GO:0008270
GO:0000166
GO:0003677
GO:0008408
GO:0005112
GO:0015930
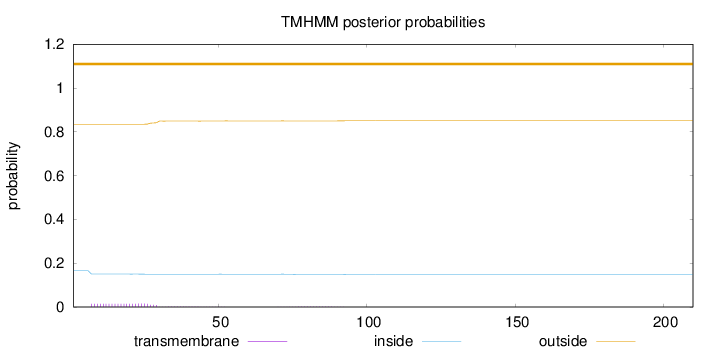
Topology
Length:
210
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.3735
Exp number, first 60 AAs:
0.34928
Total prob of N-in:
0.16519
outside
1 - 210
Population Genetic Test Statistics
Pi
294.082126
Theta
197.095842
Tajima's D
1.558594
CLR
0.736227
CSRT
0.800409979501025
Interpretation
Uncertain
