Gene
KWMTBOMO15067
Pre Gene Modal
BGIBMGA004849
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_DDB1-_and_CUL4-associated_factor_10-like_[Plutella_xylostella]
Location in the cell
Nuclear Reliability : 3.987
Sequence
CDS
ATGGGTAATCGAGATATTTTCAATTCAAGAAATTCTGCATTCAGCCCTCACAGGCTACGCGATTGGGAGATGGGATTTCAGAAGCCCATAGGGGCGGACGATGCGATGGCAAAGAGTCTGTACTGTGGTATGAAACCCATAGCTTCATGGGAATCTGACAATACAAGAACTCCTCCCACTGGAGGAGTTTTTAACCTTGAATTTTCACCTGACGGATCACTGTTAGTGGCTGCTTGTGAAAAGAAATCCTTACAAATCTTTGATCCTTTAACACACCAAAGGACACATTGTGTGACCGAAGCACATTCGGATTGTGTAAATTGTGTTAAATTCTTAGACGCTAGGATGTTTGCCACATGCTCCGATGACACAACAGTAGCACTATGGGATGTACGTAACATGAAGAAGAAGATACGTTCCCTCTTAGGTCATCTCAGATGGGTCAAAAGTATAGAATTCTCGGTTAAAGATAAATTATTAGTTACGTCTGGTTTGGATGGCAGCATATACACATGGGATATAAATTCATATACAGAATGTAATCTTGTATACCAAAGAGTATTTCATACGTCTAGTTTGATGAGGTGTCGCCTTTCGCCCGACGCAAGGAAAATGGTGCTGTGCACAACTGGAGGTTTAATAATTATTATACACGACCTTAATTTATCAACTTTAGCTCAAGATTTACATGGGTTCAAGCCGAACCTGTACAAACTGATGCAAATGAGCAAACAGCTGATACCGATAGCAGCAATGTACGATCATCTCTTTGATAGTACACGGAAGGAGAACAGAATTGAGTTTGTCTCGGACTTCCCCGACGGTAATGATGCGGAGGTCATAAGTGCATTGCAGATACACCCGCAAGGATGGAGTGCGTTAAGTCGGAATCTAAGTCACGAGGATAGGTCCGAGTGGACGTGCATTCACGACATCCAGCCGGAGGACGCGTGCCCGACGAAGCCGACGCGCGACACGAGCGCCCCCCCGCCGCGCCGCAACATGCGTCGCTTCCGTCTGCACCTGCGACTGCGACCTTCTCGGACGGAGACTGCAGCAGCTCCGAGTGAGGCGGACCCGGAGCCCGAGCCGGGCCCGTCCGGCGCCGCCGCCTCCGCCACCTCCAGGCTCTCACCGCAAGGCCTGTCGAGTATCCAAAACGACCTATGGGAGGCGTCGATAACGGTCAAACAAAACAGAATGCTTCAAGACATTTATAGCAGGGGCCAGGTACGTCGTAACTTCAACATGCAGCGGATAACCGGGATTAATACAGGGATCACTCCGCCCTCGACGCTTCGAGCGAGGCACCCAAGAGATTCCACATTTACGACGCCTTCGACGTCTGGCCACTACCCACGCGAAGTCATCATAGATAAATGCGTTTCATCGAAACCCTTCATCAGACAGAACAAGGATAGACTGCTTTACTACATTGAAGAAACGAATGAAGGCAGAGGATTCATAAAGGAATTGTGTTACTCTGCGGACGGTCGCATCGTGTGTTCGCCATTCGGGTGCGGCATGCGGCTCCTGGCTCTCAACCAGCAGTGCAGTGAGCTCCCCCACCTCGTCCAGGACTCGTCGGGGCCCAGGGTCATGACCGAAGTTGGCCAGAGTTTGGGCATTCACCAAGATCTCGTAGTTAGTTCGCGGTTCAGTCCTCGATATCACATGCTCGTCACGGGCTGTCTAGACGGCAAGATAGTCTGGTACCAGCCGTACAGCGGAGAAGATTTGTATTAA
Protein
MGNRDIFNSRNSAFSPHRLRDWEMGFQKPIGADDAMAKSLYCGMKPIASWESDNTRTPPTGGVFNLEFSPDGSLLVAACEKKSLQIFDPLTHQRTHCVTEAHSDCVNCVKFLDARMFATCSDDTTVALWDVRNMKKKIRSLLGHLRWVKSIEFSVKDKLLVTSGLDGSIYTWDINSYTECNLVYQRVFHTSSLMRCRLSPDARKMVLCTTGGLIIIIHDLNLSTLAQDLHGFKPNLYKLMQMSKQLIPIAAMYDHLFDSTRKENRIEFVSDFPDGNDAEVISALQIHPQGWSALSRNLSHEDRSEWTCIHDIQPEDACPTKPTRDTSAPPPRRNMRRFRLHLRLRPSRTETAAAPSEADPEPEPGPSGAAASATSRLSPQGLSSIQNDLWEASITVKQNRMLQDIYSRGQVRRNFNMQRITGINTGITPPSTLRARHPRDSTFTTPSTSGHYPREVIIDKCVSSKPFIRQNKDRLLYYIEETNEGRGFIKELCYSADGRIVCSPFGCGMRLLALNQQCSELPHLVQDSSGPRVMTEVGQSLGIHQDLVVSSRFSPRYHMLVTGCLDGKIVWYQPYSGEDLY
Summary
EMBL
GAMC01011246
JAB95309.1
CH479207
EDW30985.1
HM017533
ADG64906.1
+ More
HM017531 ADG64904.1 HM017525 HM017526 HM017528 HM017530 HM017534 HM017535 ADG64898.1 ADG64899.1 ADG64901.1 ADG64903.1 ADG64907.1 ADG64908.1 CH479595 EDW34941.1 HM017529 ADG64902.1 CH379063 EDY72093.2 HM017527 HM017532 HM017536 ADG64900.1 ADG64905.1 ADG64909.1 EDW34939.1
HM017531 ADG64904.1 HM017525 HM017526 HM017528 HM017530 HM017534 HM017535 ADG64898.1 ADG64899.1 ADG64901.1 ADG64903.1 ADG64907.1 ADG64908.1 CH479595 EDW34941.1 HM017529 ADG64902.1 CH379063 EDY72093.2 HM017527 HM017532 HM017536 ADG64900.1 ADG64905.1 ADG64909.1 EDW34939.1
Proteomes
Pfam
PF00400 WD40
Interpro
SUPFAM
SSF50978
SSF50978
Gene 3D
PDB
3SHF
E-value=1.12931e-11,
Score=170
Ontologies
GO
PANTHER
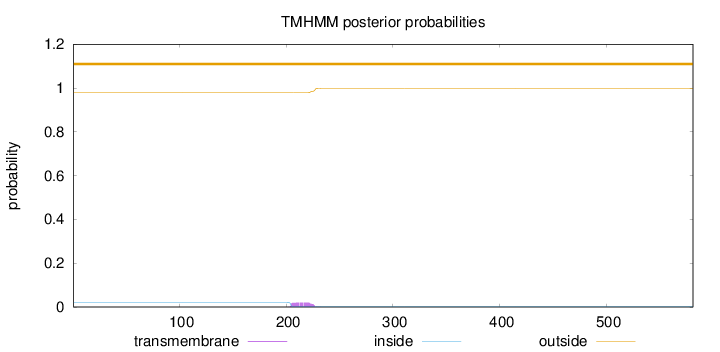
Topology
Length:
581
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.42738
Exp number, first 60 AAs:
0
Total prob of N-in:
0.02162
outside
1 - 581
Population Genetic Test Statistics
Pi
258.24602
Theta
162.28405
Tajima's D
1.438179
CLR
0.554415
CSRT
0.775211239438028
Interpretation
Uncertain
