Gene
KWMTBOMO15066 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA004824
Annotation
electron-transfer-flavoprotein_beta_polypeptide_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.829
Sequence
CDS
ATGTCTCGTGTTCTTGTTGGCGTTAAGAGAGTTATCGACTATGCTGTAAAAATTCGTGTGAAGCCAGACAAGTCTGGTGTAGTCACAGATGGGGTGAAGCACTCAATGAATCCTTTTGATGAGATAGCAGTAGAGGAGGCTGTTCGAATGAAAGAAAAAAAGTTAGCAAGCGAAGTCATTGCAGTCTCTTGTGGACCATCACAAGCACAGGAAACTCTCAGAACTGCTCTTGCGATGGGTGCAGATCGAGCTATTCATGTTGAAGTGGCTGGGGCTGAATATGACACATTACAGCCCATTCATGTTGCAAAAATCTTAGCAAAACTGTCACAAGATGAGAAGGCAGACTTAGTTATTGTTGGCAAACAGGCCATAGATGATGACTCCAACCAGACAGCACAGATGACTGCTGCTTTATTAGATTGGCCACAAGGAACATTTGCATCAAAGATAGAAAAGACTGATGGTGCTCTAACTATAACCCGTGAAATTGATGGAGGTTTAGAGGTTATCAAGACAAAAATCCCAGCTGTACTGAGTGCTGATCTGCGACTGAATGAACCACGATATGCTACACTGCCTAACATCATGAAAGCAAAGAAGAAGCCTCTCAAAAAAGTAAGTGCCAAAGACCTTGGGGTGGACCTGGCACCTCGTATCAAGGTGGTCTCTGTGGAAGATCCTCCAGTCCGACAGGCTGGCTCCATCATACCTGATGTCGACACTCTCGTTGCTAAGCTTAAGGAAGGCGGACATGTGTAA
Protein
MSRVLVGVKRVIDYAVKIRVKPDKSGVVTDGVKHSMNPFDEIAVEEAVRMKEKKLASEVIAVSCGPSQAQETLRTALAMGADRAIHVEVAGAEYDTLQPIHVAKILAKLSQDEKADLVIVGKQAIDDDSNQTAQMTAALLDWPQGTFASKIEKTDGALTITREIDGGLEVIKTKIPAVLSADLRLNEPRYATLPNIMKAKKKPLKKVSAKDLGVDLAPRIKVVSVEDPPVRQAGSIIPDVDTLVAKLKEGGHV
Summary
Uniprot
Q2F6A1
A0A2A4JMV5
A0A212FLM7
A0A3S2NLH6
A0A194PM61
S4NJ39
+ More
A0A2H1WV21 A0A0N1I7H4 A0A1B6F3Z7 A0A1B6EF91 A0A2J7QLI2 K7IR93 D6WSM6 W8CCA7 A0A195FPB7 A0A1Y1MB95 A0A0T6ASW5 A0A151WM30 E2AIR9 A0A0J7L4M3 A0A1A9VDP3 A0A182G5Z7 A0A026WVS2 A0A1W4WX89 F4WJ49 A0A023ENM9 B0W9P9 A0A0K8VV21 T1PHZ9 J3JYG8 N6U2K5 A0A034VPU8 A0A0C9R912 A0A1L8EHB8 A0A1A9ZSV8 D3TNF0 A0A0A1XT47 A0A1A9YQ62 A0A195BMF2 A0A1L8E115 A0A158NTV2 A0A1A9W5P6 E2B6H4 A0A151JR04 A0A1B0C9H6 A0A182ITB0 A0A067RME8 A0A1I8PJD8 A0A182Q436 B4PPJ8 B3P5C9 A0A182NN50 A0A0K8TSS5 A0A195CTV2 A0A0C9QL89 Q8MR58 B4R0S9 B4HZF2 Q0KHZ6 A0A1B0DM44 W5JJZ6 B3LWX6 A0A084W805 A0A0A9XI89 A0A0M5JCP3 A0A182PST4 Q297T4 A0A182LQ71 Q7Q501 A0A182I9E3 A0A182X4S9 A0A182V5E2 A0A182RE72 A0A182YEP7 A0A182TDY7 A0A182T8P6 B4KAN5 A0A1W4UC91 A0A2M3Z262 B4G306 A0A2M4A8G6 A0A3B0JF93 A0A0M8ZSX3 A0A182LW32 T1DNI8 A0A2M4BYC4 A0A0L0BYC8 A0A0P4VLG6 T1IBK0 A0A310SLQ8 A0A182VPX5 A0A182F8S1 R4FQ22 A0A0L7R2V3 A0A088A7F1 U4UD08 A0A2A3EAZ4 U4UF37 A0A069DRJ2 U5EYT1
A0A2H1WV21 A0A0N1I7H4 A0A1B6F3Z7 A0A1B6EF91 A0A2J7QLI2 K7IR93 D6WSM6 W8CCA7 A0A195FPB7 A0A1Y1MB95 A0A0T6ASW5 A0A151WM30 E2AIR9 A0A0J7L4M3 A0A1A9VDP3 A0A182G5Z7 A0A026WVS2 A0A1W4WX89 F4WJ49 A0A023ENM9 B0W9P9 A0A0K8VV21 T1PHZ9 J3JYG8 N6U2K5 A0A034VPU8 A0A0C9R912 A0A1L8EHB8 A0A1A9ZSV8 D3TNF0 A0A0A1XT47 A0A1A9YQ62 A0A195BMF2 A0A1L8E115 A0A158NTV2 A0A1A9W5P6 E2B6H4 A0A151JR04 A0A1B0C9H6 A0A182ITB0 A0A067RME8 A0A1I8PJD8 A0A182Q436 B4PPJ8 B3P5C9 A0A182NN50 A0A0K8TSS5 A0A195CTV2 A0A0C9QL89 Q8MR58 B4R0S9 B4HZF2 Q0KHZ6 A0A1B0DM44 W5JJZ6 B3LWX6 A0A084W805 A0A0A9XI89 A0A0M5JCP3 A0A182PST4 Q297T4 A0A182LQ71 Q7Q501 A0A182I9E3 A0A182X4S9 A0A182V5E2 A0A182RE72 A0A182YEP7 A0A182TDY7 A0A182T8P6 B4KAN5 A0A1W4UC91 A0A2M3Z262 B4G306 A0A2M4A8G6 A0A3B0JF93 A0A0M8ZSX3 A0A182LW32 T1DNI8 A0A2M4BYC4 A0A0L0BYC8 A0A0P4VLG6 T1IBK0 A0A310SLQ8 A0A182VPX5 A0A182F8S1 R4FQ22 A0A0L7R2V3 A0A088A7F1 U4UD08 A0A2A3EAZ4 U4UF37 A0A069DRJ2 U5EYT1
Pubmed
19121390
22118469
26354079
23622113
20075255
18362917
+ More
19820115 24495485 28004739 20798317 26483478 24508170 30249741 21719571 24945155 25315136 22516182 23537049 25348373 20353571 25830018 21347285 24845553 17994087 17550304 26369729 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20920257 23761445 18057021 24438588 25401762 26823975 15632085 23185243 20966253 12364791 14747013 17210077 25244985 26108605 27129103 26334808
19820115 24495485 28004739 20798317 26483478 24508170 30249741 21719571 24945155 25315136 22516182 23537049 25348373 20353571 25830018 21347285 24845553 17994087 17550304 26369729 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20920257 23761445 18057021 24438588 25401762 26823975 15632085 23185243 20966253 12364791 14747013 17210077 25244985 26108605 27129103 26334808
EMBL
BABH01025030
DQ311171
ABD36116.1
NWSH01000949
PCG73405.1
AGBW02007763
+ More
OWR54600.1 RSAL01000008 RVE53944.1 KQ459604 KPI92200.1 GAIX01013834 JAA78726.1 ODYU01011277 SOQ56915.1 KQ460594 KPJ13923.1 GECZ01025176 JAS44593.1 GEDC01000709 JAS36589.1 NEVH01013243 PNF29454.1 KQ971354 EFA05895.1 GAMC01004026 JAC02530.1 KQ981382 KYN42303.1 GEZM01035760 JAV83099.1 LJIG01022885 KRT78227.1 KQ982959 KYQ48755.1 GL439869 EFN66681.1 LBMM01000651 KMQ97887.1 JXUM01042943 JXUM01044222 KQ561391 KQ561344 KXJ78732.1 KXJ78884.1 KK107079 QOIP01000010 EZA60130.1 RLU17817.1 GL888181 EGI65819.1 GAPW01003569 JAC10029.1 DS231865 EDS40368.1 GDHF01009587 GDHF01003129 JAI42727.1 JAI49185.1 KA647508 AFP62137.1 BT128296 AEE63254.1 APGK01052246 KB741211 ENN72797.1 GAKP01013636 JAC45316.1 GBYB01004650 JAG74417.1 GFDG01000754 JAV18045.1 CCAG010010757 EZ422952 ADD19228.1 GBXI01000061 JAD14231.1 KQ976435 KYM87055.1 GFDF01001748 JAV12336.1 ADTU01026166 GL445973 EFN88718.1 KQ978615 KYN29828.1 AJWK01002349 KK852540 KDR21775.1 AXCN02002214 CM000160 EDW98248.1 CH954182 EDV53179.1 GDAI01000179 JAI17424.1 KQ977279 KYN04095.1 GBYB01001352 JAG71119.1 AY122112 AAM52624.1 CM000364 EDX14900.1 CH480819 EDW53409.1 AE014297 AAF56940.1 AAN14188.1 AJVK01016664 ADMH02001214 ETN63633.1 CH902617 EDV43815.1 KPU80482.1 ATLV01021336 KE525316 KFB46349.1 GBHO01025086 GBHO01025085 GBRD01007754 GDHC01014085 GDHC01001019 JAG18518.1 JAG18519.1 JAG58067.1 JAQ04544.1 JAQ17610.1 CP012526 ALC46918.1 CM000070 EAL28121.1 KRT00471.1 KRT00472.1 KRT00473.1 KRT00474.1 AAAB01008961 EAA11983.3 APCN01008266 CH933806 EDW16772.2 GGFM01001840 MBW22591.1 CH479179 EDW24201.1 GGFK01003773 MBW37094.1 OUUW01000005 SPP81054.1 KQ435866 KOX70297.1 AXCM01000327 GAMD01002830 JAA98760.1 GGFJ01008597 MBW57738.1 JRES01001160 KNC25043.1 GDKW01000639 JAI55956.1 ACPB03006483 KQ762343 OAD55932.1 GAHY01000629 JAA76881.1 KQ414664 KOC65207.1 KB632263 ERL90932.1 KZ288296 PBC28943.1 ERL90933.1 GBGD01002627 JAC86262.1 GANO01001831 JAB58040.1
OWR54600.1 RSAL01000008 RVE53944.1 KQ459604 KPI92200.1 GAIX01013834 JAA78726.1 ODYU01011277 SOQ56915.1 KQ460594 KPJ13923.1 GECZ01025176 JAS44593.1 GEDC01000709 JAS36589.1 NEVH01013243 PNF29454.1 KQ971354 EFA05895.1 GAMC01004026 JAC02530.1 KQ981382 KYN42303.1 GEZM01035760 JAV83099.1 LJIG01022885 KRT78227.1 KQ982959 KYQ48755.1 GL439869 EFN66681.1 LBMM01000651 KMQ97887.1 JXUM01042943 JXUM01044222 KQ561391 KQ561344 KXJ78732.1 KXJ78884.1 KK107079 QOIP01000010 EZA60130.1 RLU17817.1 GL888181 EGI65819.1 GAPW01003569 JAC10029.1 DS231865 EDS40368.1 GDHF01009587 GDHF01003129 JAI42727.1 JAI49185.1 KA647508 AFP62137.1 BT128296 AEE63254.1 APGK01052246 KB741211 ENN72797.1 GAKP01013636 JAC45316.1 GBYB01004650 JAG74417.1 GFDG01000754 JAV18045.1 CCAG010010757 EZ422952 ADD19228.1 GBXI01000061 JAD14231.1 KQ976435 KYM87055.1 GFDF01001748 JAV12336.1 ADTU01026166 GL445973 EFN88718.1 KQ978615 KYN29828.1 AJWK01002349 KK852540 KDR21775.1 AXCN02002214 CM000160 EDW98248.1 CH954182 EDV53179.1 GDAI01000179 JAI17424.1 KQ977279 KYN04095.1 GBYB01001352 JAG71119.1 AY122112 AAM52624.1 CM000364 EDX14900.1 CH480819 EDW53409.1 AE014297 AAF56940.1 AAN14188.1 AJVK01016664 ADMH02001214 ETN63633.1 CH902617 EDV43815.1 KPU80482.1 ATLV01021336 KE525316 KFB46349.1 GBHO01025086 GBHO01025085 GBRD01007754 GDHC01014085 GDHC01001019 JAG18518.1 JAG18519.1 JAG58067.1 JAQ04544.1 JAQ17610.1 CP012526 ALC46918.1 CM000070 EAL28121.1 KRT00471.1 KRT00472.1 KRT00473.1 KRT00474.1 AAAB01008961 EAA11983.3 APCN01008266 CH933806 EDW16772.2 GGFM01001840 MBW22591.1 CH479179 EDW24201.1 GGFK01003773 MBW37094.1 OUUW01000005 SPP81054.1 KQ435866 KOX70297.1 AXCM01000327 GAMD01002830 JAA98760.1 GGFJ01008597 MBW57738.1 JRES01001160 KNC25043.1 GDKW01000639 JAI55956.1 ACPB03006483 KQ762343 OAD55932.1 GAHY01000629 JAA76881.1 KQ414664 KOC65207.1 KB632263 ERL90932.1 KZ288296 PBC28943.1 ERL90933.1 GBGD01002627 JAC86262.1 GANO01001831 JAB58040.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000283053
UP000053268
UP000053240
+ More
UP000235965 UP000002358 UP000007266 UP000078541 UP000075809 UP000000311 UP000036403 UP000078200 UP000069940 UP000249989 UP000053097 UP000279307 UP000192223 UP000007755 UP000002320 UP000095301 UP000019118 UP000092445 UP000092444 UP000092443 UP000078540 UP000005205 UP000091820 UP000008237 UP000078492 UP000092461 UP000075880 UP000027135 UP000095300 UP000075886 UP000002282 UP000008711 UP000075884 UP000078542 UP000000304 UP000001292 UP000000803 UP000092462 UP000000673 UP000007801 UP000030765 UP000092553 UP000075885 UP000001819 UP000075882 UP000007062 UP000075840 UP000076407 UP000075903 UP000075900 UP000076408 UP000075902 UP000075901 UP000009192 UP000192221 UP000008744 UP000268350 UP000053105 UP000075883 UP000037069 UP000015103 UP000075920 UP000069272 UP000053825 UP000005203 UP000030742 UP000242457
UP000235965 UP000002358 UP000007266 UP000078541 UP000075809 UP000000311 UP000036403 UP000078200 UP000069940 UP000249989 UP000053097 UP000279307 UP000192223 UP000007755 UP000002320 UP000095301 UP000019118 UP000092445 UP000092444 UP000092443 UP000078540 UP000005205 UP000091820 UP000008237 UP000078492 UP000092461 UP000075880 UP000027135 UP000095300 UP000075886 UP000002282 UP000008711 UP000075884 UP000078542 UP000000304 UP000001292 UP000000803 UP000092462 UP000000673 UP000007801 UP000030765 UP000092553 UP000075885 UP000001819 UP000075882 UP000007062 UP000075840 UP000076407 UP000075903 UP000075900 UP000076408 UP000075902 UP000075901 UP000009192 UP000192221 UP000008744 UP000268350 UP000053105 UP000075883 UP000037069 UP000015103 UP000075920 UP000069272 UP000053825 UP000005203 UP000030742 UP000242457
PRIDE
Interpro
SUPFAM
SSF46785
SSF46785
Gene 3D
CDD
ProteinModelPortal
Q2F6A1
A0A2A4JMV5
A0A212FLM7
A0A3S2NLH6
A0A194PM61
S4NJ39
+ More
A0A2H1WV21 A0A0N1I7H4 A0A1B6F3Z7 A0A1B6EF91 A0A2J7QLI2 K7IR93 D6WSM6 W8CCA7 A0A195FPB7 A0A1Y1MB95 A0A0T6ASW5 A0A151WM30 E2AIR9 A0A0J7L4M3 A0A1A9VDP3 A0A182G5Z7 A0A026WVS2 A0A1W4WX89 F4WJ49 A0A023ENM9 B0W9P9 A0A0K8VV21 T1PHZ9 J3JYG8 N6U2K5 A0A034VPU8 A0A0C9R912 A0A1L8EHB8 A0A1A9ZSV8 D3TNF0 A0A0A1XT47 A0A1A9YQ62 A0A195BMF2 A0A1L8E115 A0A158NTV2 A0A1A9W5P6 E2B6H4 A0A151JR04 A0A1B0C9H6 A0A182ITB0 A0A067RME8 A0A1I8PJD8 A0A182Q436 B4PPJ8 B3P5C9 A0A182NN50 A0A0K8TSS5 A0A195CTV2 A0A0C9QL89 Q8MR58 B4R0S9 B4HZF2 Q0KHZ6 A0A1B0DM44 W5JJZ6 B3LWX6 A0A084W805 A0A0A9XI89 A0A0M5JCP3 A0A182PST4 Q297T4 A0A182LQ71 Q7Q501 A0A182I9E3 A0A182X4S9 A0A182V5E2 A0A182RE72 A0A182YEP7 A0A182TDY7 A0A182T8P6 B4KAN5 A0A1W4UC91 A0A2M3Z262 B4G306 A0A2M4A8G6 A0A3B0JF93 A0A0M8ZSX3 A0A182LW32 T1DNI8 A0A2M4BYC4 A0A0L0BYC8 A0A0P4VLG6 T1IBK0 A0A310SLQ8 A0A182VPX5 A0A182F8S1 R4FQ22 A0A0L7R2V3 A0A088A7F1 U4UD08 A0A2A3EAZ4 U4UF37 A0A069DRJ2 U5EYT1
A0A2H1WV21 A0A0N1I7H4 A0A1B6F3Z7 A0A1B6EF91 A0A2J7QLI2 K7IR93 D6WSM6 W8CCA7 A0A195FPB7 A0A1Y1MB95 A0A0T6ASW5 A0A151WM30 E2AIR9 A0A0J7L4M3 A0A1A9VDP3 A0A182G5Z7 A0A026WVS2 A0A1W4WX89 F4WJ49 A0A023ENM9 B0W9P9 A0A0K8VV21 T1PHZ9 J3JYG8 N6U2K5 A0A034VPU8 A0A0C9R912 A0A1L8EHB8 A0A1A9ZSV8 D3TNF0 A0A0A1XT47 A0A1A9YQ62 A0A195BMF2 A0A1L8E115 A0A158NTV2 A0A1A9W5P6 E2B6H4 A0A151JR04 A0A1B0C9H6 A0A182ITB0 A0A067RME8 A0A1I8PJD8 A0A182Q436 B4PPJ8 B3P5C9 A0A182NN50 A0A0K8TSS5 A0A195CTV2 A0A0C9QL89 Q8MR58 B4R0S9 B4HZF2 Q0KHZ6 A0A1B0DM44 W5JJZ6 B3LWX6 A0A084W805 A0A0A9XI89 A0A0M5JCP3 A0A182PST4 Q297T4 A0A182LQ71 Q7Q501 A0A182I9E3 A0A182X4S9 A0A182V5E2 A0A182RE72 A0A182YEP7 A0A182TDY7 A0A182T8P6 B4KAN5 A0A1W4UC91 A0A2M3Z262 B4G306 A0A2M4A8G6 A0A3B0JF93 A0A0M8ZSX3 A0A182LW32 T1DNI8 A0A2M4BYC4 A0A0L0BYC8 A0A0P4VLG6 T1IBK0 A0A310SLQ8 A0A182VPX5 A0A182F8S1 R4FQ22 A0A0L7R2V3 A0A088A7F1 U4UD08 A0A2A3EAZ4 U4UF37 A0A069DRJ2 U5EYT1
PDB
1T9G
E-value=1.58624e-84,
Score=794
Ontologies
GO
PANTHER
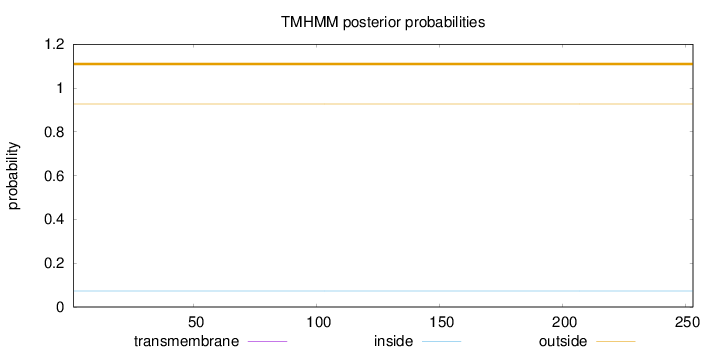
Topology
Length:
253
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0002
Exp number, first 60 AAs:
6e-05
Total prob of N-in:
0.07313
outside
1 - 253
Population Genetic Test Statistics
Pi
378.01387
Theta
233.358103
Tajima's D
1.952297
CLR
0.000517
CSRT
0.874906254687266
Interpretation
Uncertain
