Gene
KWMTBOMO15063 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA004822
Annotation
eukaryotic_initiation_factor_4A-III_[Bombyx_mori]
Full name
Eukaryotic initiation factor 4A-III
Location in the cell
Nuclear Reliability : 3.172
Sequence
CDS
ATGACATCGTCTGAAGTATCGTCCAATAGAAAAATATTGTCGGAAGATTTGTCGAATGTTGAGTTCGACACGAGTGAAGATGTCGAAGTTATTCCCACTTTTGACTCCATGGGACTTCGAGATGAGTTATTGCGAGGAATATACACATATGGTTTCGAAAAACCTTCAGCAATTCAGCAGAGAAGCATCTTGCCTATAGTCAAAGGCAGAGATGTAATAGCTCAAGCACAATCTGGTACAGGAAAGACTGCGACATTCTCAATTTCAATACTACAGACTTTAGACACGACTCTGCGTGAAACTCAAGTACTTATTCTTTCACCGACCCGTGAACTGGCTACTCAGATTCAGAAAGTGATATTAGCTCTTGGAGATTTCATGAACGTTCAATGTCATGCTTGTATTGGAGGTACCAATCTTGGAGAAGATATAAGGAAATTGGATTACGGACAGCATGTTGTGTCTGGCACACCTGGCAGAGTTTTTGATATGATCAGAAGGCGTGTGTTGAGAACCCGGTCAATAAAGATGCTGGTTCTTGATGAGGCTGATGAGATGTTGAACAAAGGTTTCAAAGAGCAAATTTATGATGTATACCGCTATCTGCCTCCTGCAACTCAGGTTGTGCTAATATCAGCAACACTACCCCATGAGATATTGGAAATGACATCAAAGTTTATGACTGATCCAATAAGGATATTAGTAAAACGTGATGAGTTGACACTGGAGGGTATCAAACAGTTCTTCGTGGCAGTTGAACGAGAAGAATGGAAGTTTGACACACTTTGTGACCTCTATGATACACTGACAATTACCCAAGCAGTAATATTTTGTAATACAAAGAGGAAGGTTGACTGGCTCACACAGAAGATGCAAGAGGCAAACTTCACTGTCAGTTCAATGCATGGTGACATGCCACAGAAAGAGAGAGACAACATTATGAAGGAATTTCGTTCTGGTCAAAGCCGTGTGCTCATTACAACTGACGTCTGGGCACGAGGCATTGATGTCCAACAAGTATCCTTGGTTATAAACTATGACTTGCCCAACAATCGTGAGTTATACATTCACAGAATTGGTAGATCGGGTCGATTCGGTCGTAAGGGTGTCGCCATCAACTTCGTTAAATCAGATGACATCAGGATCTTGAGAGACATTGAACAGTACTACTCTACTCAAATAGATGAAATGCCAATGAATGTGGCTGACTTAATATAA
Protein
MTSSEVSSNRKILSEDLSNVEFDTSEDVEVIPTFDSMGLRDELLRGIYTYGFEKPSAIQQRSILPIVKGRDVIAQAQSGTGKTATFSISILQTLDTTLRETQVLILSPTRELATQIQKVILALGDFMNVQCHACIGGTNLGEDIRKLDYGQHVVSGTPGRVFDMIRRRVLRTRSIKMLVLDEADEMLNKGFKEQIYDVYRYLPPATQVVLISATLPHEILEMTSKFMTDPIRILVKRDELTLEGIKQFFVAVEREEWKFDTLCDLYDTLTITQAVIFCNTKRKVDWLTQKMQEANFTVSSMHGDMPQKERDNIMKEFRSGQSRVLITTDVWARGIDVQQVSLVINYDLPNNRELYIHRIGRSGRFGRKGVAINFVKSDDIRILRDIEQYYSTQIDEMPMNVADLI
Summary
Description
ATP-dependent RNA helicase (PubMed:22961380). Core component of the splicing-dependent multiprotein exon junction complex (EJC) deposited at splice junctions on mRNAs (PubMed:14973490, PubMed:22961380). Involved in exon definition of genes containing long introns, including the rolled/MAPK gene (PubMed:20946982, PubMed:20946983). Has a role in oskar mRNA localization at the posterior pole of the developing oocyte (PubMed:14973490).
Catalytic Activity
ATP + H2O = ADP + H(+) + phosphate
Subunit
Part of the mRNA splicing-dependent exon junction complex (EJC) complex; the core complex contains btz/CASC3, eIF4AIII, mago and tsu/RBM8A (PubMed:14973490, PubMed:22961380). Interacts with btz/CASC3 and mago (PubMed:14973490). Interacts with ncm/CWC22 (PubMed:22961380).
Similarity
Belongs to the DEAD box helicase family.
Keywords
ATP-binding
Complete proteome
Developmental protein
Helicase
Hydrolase
mRNA processing
mRNA splicing
mRNA transport
Nucleotide-binding
Nucleus
Reference proteome
RNA-binding
Transport
Feature
chain Eukaryotic initiation factor 4A-III
Uniprot
Q0PZE7
A0A212FLI2
A0A2W1BRX8
A0A2A4JP97
A0A3S2PKD7
A0A2H1WTH0
+ More
I4DME1 I4DJR8 A0A194PFX7 A0A0N1PHX3 S4PWN2 A0A1Q3FV91 A0A2J7QNB4 A0A0M8ZSQ6 A0A067RMC6 A0A1B0CXD4 A0A154P503 A0A182J1G5 A0A2A3ERN3 A0A087ZZS7 A0A0L7R1R2 E2BEH0 A0A182QXS5 A0A182N6M7 A0A084W917 Q16GE2 A0A182VJT6 A0A1B0DM43 A0A182P301 A0A182MFM2 A0A182SNI4 A0A182UES5 A0A182X8D7 Q7PQQ2 A0A182RXB9 A0A182HIZ1 A0A1L8DGW1 A0A0J7KKY9 E9IZK2 A0A2M3Z6Y1 A0A2M4AH50 A0A2M4BQA4 W5JLN7 A0A151J4R6 A0A182XVP1 A0A151JVP6 A0A182JWR5 A0A026WX81 A0A195CSB2 A0A158NIN0 E2AWS1 A0A0K8TMU1 A0A1L8DGS9 A0A1B6L3M7 A0A232ELK5 W8BID8 A0A1B6FKM8 A0A1B6IUQ3 B4NGN0 A0A1W4VH90 B4LYN9 B4K8T2 B4PT42 A0A0M3QX77 B3M0D8 B4JV86 B3NZU1 Q9VHS8 A0A0K8UBI3 A0A034VAP8 A0A1B6C2G1 A0A0L0CBE0 B4HLL9 A0A3B0JQK9 T1PG03 Q298G7 B4G3R2 A0A1I8Q8H2 A0A1A9WML1 A0A1B0FFV4 A0A1A9YQ17 A0A1A9UFH7 A0A1B0B4Y7 A0A1A9ZT23 E0W2V0 A0A3S3SQE4 A0A1S3J3P5 T1K100 A0A2S1TJI0 A0A0K2T8I3 G3MPX6 A0A023GJK7 A0A0P5URV6 A0A0P6BGL6 T1E1I3 L7M7P7 A0A131XB35 A0A0P5CLK1 A0A0P5VWE4 C1C2L4
I4DME1 I4DJR8 A0A194PFX7 A0A0N1PHX3 S4PWN2 A0A1Q3FV91 A0A2J7QNB4 A0A0M8ZSQ6 A0A067RMC6 A0A1B0CXD4 A0A154P503 A0A182J1G5 A0A2A3ERN3 A0A087ZZS7 A0A0L7R1R2 E2BEH0 A0A182QXS5 A0A182N6M7 A0A084W917 Q16GE2 A0A182VJT6 A0A1B0DM43 A0A182P301 A0A182MFM2 A0A182SNI4 A0A182UES5 A0A182X8D7 Q7PQQ2 A0A182RXB9 A0A182HIZ1 A0A1L8DGW1 A0A0J7KKY9 E9IZK2 A0A2M3Z6Y1 A0A2M4AH50 A0A2M4BQA4 W5JLN7 A0A151J4R6 A0A182XVP1 A0A151JVP6 A0A182JWR5 A0A026WX81 A0A195CSB2 A0A158NIN0 E2AWS1 A0A0K8TMU1 A0A1L8DGS9 A0A1B6L3M7 A0A232ELK5 W8BID8 A0A1B6FKM8 A0A1B6IUQ3 B4NGN0 A0A1W4VH90 B4LYN9 B4K8T2 B4PT42 A0A0M3QX77 B3M0D8 B4JV86 B3NZU1 Q9VHS8 A0A0K8UBI3 A0A034VAP8 A0A1B6C2G1 A0A0L0CBE0 B4HLL9 A0A3B0JQK9 T1PG03 Q298G7 B4G3R2 A0A1I8Q8H2 A0A1A9WML1 A0A1B0FFV4 A0A1A9YQ17 A0A1A9UFH7 A0A1B0B4Y7 A0A1A9ZT23 E0W2V0 A0A3S3SQE4 A0A1S3J3P5 T1K100 A0A2S1TJI0 A0A0K2T8I3 G3MPX6 A0A023GJK7 A0A0P5URV6 A0A0P6BGL6 T1E1I3 L7M7P7 A0A131XB35 A0A0P5CLK1 A0A0P5VWE4 C1C2L4
EC Number
3.6.4.13
Pubmed
19121390
22118469
28756777
22651552
26354079
23622113
+ More
24845553 20798317 24438588 17510324 12364791 14747013 17210077 21282665 20920257 23761445 25244985 24508170 30249741 21347285 26369729 28648823 24495485 17994087 17550304 10731132 12537572 14973490 20946982 20946983 22961380 25348373 26108605 25315136 15632085 20566863 22216098 24330624 25576852 28049606
24845553 20798317 24438588 17510324 12364791 14747013 17210077 21282665 20920257 23761445 25244985 24508170 30249741 21347285 26369729 28648823 24495485 17994087 17550304 10731132 12537572 14973490 20946982 20946983 22961380 25348373 26108605 25315136 15632085 20566863 22216098 24330624 25576852 28049606
EMBL
BABH01025034
DQ667967
ABG73410.1
AGBW02007763
OWR54603.1
KZ149918
+ More
PZC77842.1 NWSH01000949 PCG73414.1 RSAL01000008 RVE53942.1 ODYU01010900 SOQ56297.1 AK402459 BAM19081.1 AK401536 BAM18158.1 KQ459604 KPI92197.1 KQ460594 KPJ13926.1 GAIX01006493 JAA86067.1 GFDL01003531 JAV31514.1 NEVH01013196 PNF30072.1 KQ435863 KOX70370.1 KK852425 KDR24148.1 AJWK01033613 KQ434809 KZC06210.1 KZ288194 PBC33932.1 KQ414667 KOC64731.1 GL447812 EFN85902.1 AXCN02000299 ATLV01021572 KE525320 KFB46711.1 CH478287 EAT33304.1 AJVK01016664 AXCM01000558 AAAB01008879 EAA08469.4 APCN01002105 GFDF01008423 JAV05661.1 LBMM01006135 KMQ90891.1 GL767232 EFZ14001.1 GGFM01003447 MBW24198.1 GGFK01006721 MBW40042.1 GGFJ01006061 MBW55202.1 ADMH02001068 ETN64218.1 KQ980102 KYN17746.1 KQ981702 KYN37460.1 KK107072 QOIP01000007 EZA60660.1 RLU20877.1 KQ977306 KYN03623.1 ADTU01017000 GL443424 EFN62107.1 GDAI01001931 JAI15672.1 GFDF01008422 JAV05662.1 GEBQ01021736 JAT18241.1 NNAY01003564 OXU19192.1 GAMC01005535 JAC01021.1 GECZ01019216 JAS50553.1 GECU01017032 JAS90674.1 CH964272 EDW84377.2 CH940650 EDW68059.1 CH933806 EDW15501.1 CM000160 EDW96503.1 CP012526 ALC45453.1 CH902617 EDV43144.1 CH916374 EDV91406.1 CH954181 EDV49939.1 AE014297 AY089635 BT133445 AAF54221.1 AAL90373.1 AFH89818.1 GDHF01028310 JAI24004.1 GAKP01019785 JAC39167.1 GEDC01029878 JAS07420.1 JRES01000634 KNC29728.1 CH480815 EDW43046.1 OUUW01000007 SPP82662.1 KA647075 AFP61704.1 CM000070 EAL27988.1 CH479179 EDW24383.1 CCAG010004727 JXJN01008574 DS235879 EEB19956.1 NCKU01000061 RWS17558.1 CAEY01001145 CAEY01001146 KY982623 AWI47734.1 HACA01005023 CDW22384.1 JO843927 AEO35544.1 GBBM01001292 GBBM01001291 JAC34127.1 GDIP01111056 JAL92658.1 GDIP01198200 GDIP01197293 GDIP01181843 GDIP01029558 JAM74157.1 GALA01001639 JAA93213.1 GACK01004708 JAA60326.1 GEFH01004268 JAP64313.1 GDIP01168739 JAJ54663.1 GDIP01200627 GDIP01198199 GDIP01196289 GDIP01195089 GDIP01183393 GDIP01181842 GDIP01111055 GDIP01094302 GDIP01094301 GDIP01086556 GDIP01052356 JAM09413.1 BT081093 ACO15517.1
PZC77842.1 NWSH01000949 PCG73414.1 RSAL01000008 RVE53942.1 ODYU01010900 SOQ56297.1 AK402459 BAM19081.1 AK401536 BAM18158.1 KQ459604 KPI92197.1 KQ460594 KPJ13926.1 GAIX01006493 JAA86067.1 GFDL01003531 JAV31514.1 NEVH01013196 PNF30072.1 KQ435863 KOX70370.1 KK852425 KDR24148.1 AJWK01033613 KQ434809 KZC06210.1 KZ288194 PBC33932.1 KQ414667 KOC64731.1 GL447812 EFN85902.1 AXCN02000299 ATLV01021572 KE525320 KFB46711.1 CH478287 EAT33304.1 AJVK01016664 AXCM01000558 AAAB01008879 EAA08469.4 APCN01002105 GFDF01008423 JAV05661.1 LBMM01006135 KMQ90891.1 GL767232 EFZ14001.1 GGFM01003447 MBW24198.1 GGFK01006721 MBW40042.1 GGFJ01006061 MBW55202.1 ADMH02001068 ETN64218.1 KQ980102 KYN17746.1 KQ981702 KYN37460.1 KK107072 QOIP01000007 EZA60660.1 RLU20877.1 KQ977306 KYN03623.1 ADTU01017000 GL443424 EFN62107.1 GDAI01001931 JAI15672.1 GFDF01008422 JAV05662.1 GEBQ01021736 JAT18241.1 NNAY01003564 OXU19192.1 GAMC01005535 JAC01021.1 GECZ01019216 JAS50553.1 GECU01017032 JAS90674.1 CH964272 EDW84377.2 CH940650 EDW68059.1 CH933806 EDW15501.1 CM000160 EDW96503.1 CP012526 ALC45453.1 CH902617 EDV43144.1 CH916374 EDV91406.1 CH954181 EDV49939.1 AE014297 AY089635 BT133445 AAF54221.1 AAL90373.1 AFH89818.1 GDHF01028310 JAI24004.1 GAKP01019785 JAC39167.1 GEDC01029878 JAS07420.1 JRES01000634 KNC29728.1 CH480815 EDW43046.1 OUUW01000007 SPP82662.1 KA647075 AFP61704.1 CM000070 EAL27988.1 CH479179 EDW24383.1 CCAG010004727 JXJN01008574 DS235879 EEB19956.1 NCKU01000061 RWS17558.1 CAEY01001145 CAEY01001146 KY982623 AWI47734.1 HACA01005023 CDW22384.1 JO843927 AEO35544.1 GBBM01001292 GBBM01001291 JAC34127.1 GDIP01111056 JAL92658.1 GDIP01198200 GDIP01197293 GDIP01181843 GDIP01029558 JAM74157.1 GALA01001639 JAA93213.1 GACK01004708 JAA60326.1 GEFH01004268 JAP64313.1 GDIP01168739 JAJ54663.1 GDIP01200627 GDIP01198199 GDIP01196289 GDIP01195089 GDIP01183393 GDIP01181842 GDIP01111055 GDIP01094302 GDIP01094301 GDIP01086556 GDIP01052356 JAM09413.1 BT081093 ACO15517.1
Proteomes
UP000005204
UP000007151
UP000218220
UP000283053
UP000053268
UP000053240
+ More
UP000235965 UP000053105 UP000027135 UP000092461 UP000076502 UP000075880 UP000242457 UP000005203 UP000053825 UP000008237 UP000075886 UP000075884 UP000030765 UP000008820 UP000075903 UP000092462 UP000075885 UP000075883 UP000075901 UP000075902 UP000076407 UP000007062 UP000075900 UP000075840 UP000036403 UP000000673 UP000078492 UP000076408 UP000078541 UP000075881 UP000053097 UP000279307 UP000078542 UP000005205 UP000000311 UP000215335 UP000007798 UP000192221 UP000008792 UP000009192 UP000002282 UP000092553 UP000007801 UP000001070 UP000008711 UP000000803 UP000037069 UP000001292 UP000268350 UP000095301 UP000001819 UP000008744 UP000095300 UP000091820 UP000092444 UP000092443 UP000078200 UP000092460 UP000092445 UP000009046 UP000285301 UP000085678 UP000015104
UP000235965 UP000053105 UP000027135 UP000092461 UP000076502 UP000075880 UP000242457 UP000005203 UP000053825 UP000008237 UP000075886 UP000075884 UP000030765 UP000008820 UP000075903 UP000092462 UP000075885 UP000075883 UP000075901 UP000075902 UP000076407 UP000007062 UP000075900 UP000075840 UP000036403 UP000000673 UP000078492 UP000076408 UP000078541 UP000075881 UP000053097 UP000279307 UP000078542 UP000005205 UP000000311 UP000215335 UP000007798 UP000192221 UP000008792 UP000009192 UP000002282 UP000092553 UP000007801 UP000001070 UP000008711 UP000000803 UP000037069 UP000001292 UP000268350 UP000095301 UP000001819 UP000008744 UP000095300 UP000091820 UP000092444 UP000092443 UP000078200 UP000092460 UP000092445 UP000009046 UP000285301 UP000085678 UP000015104
Interpro
SUPFAM
SSF52540
SSF52540
CDD
ProteinModelPortal
Q0PZE7
A0A212FLI2
A0A2W1BRX8
A0A2A4JP97
A0A3S2PKD7
A0A2H1WTH0
+ More
I4DME1 I4DJR8 A0A194PFX7 A0A0N1PHX3 S4PWN2 A0A1Q3FV91 A0A2J7QNB4 A0A0M8ZSQ6 A0A067RMC6 A0A1B0CXD4 A0A154P503 A0A182J1G5 A0A2A3ERN3 A0A087ZZS7 A0A0L7R1R2 E2BEH0 A0A182QXS5 A0A182N6M7 A0A084W917 Q16GE2 A0A182VJT6 A0A1B0DM43 A0A182P301 A0A182MFM2 A0A182SNI4 A0A182UES5 A0A182X8D7 Q7PQQ2 A0A182RXB9 A0A182HIZ1 A0A1L8DGW1 A0A0J7KKY9 E9IZK2 A0A2M3Z6Y1 A0A2M4AH50 A0A2M4BQA4 W5JLN7 A0A151J4R6 A0A182XVP1 A0A151JVP6 A0A182JWR5 A0A026WX81 A0A195CSB2 A0A158NIN0 E2AWS1 A0A0K8TMU1 A0A1L8DGS9 A0A1B6L3M7 A0A232ELK5 W8BID8 A0A1B6FKM8 A0A1B6IUQ3 B4NGN0 A0A1W4VH90 B4LYN9 B4K8T2 B4PT42 A0A0M3QX77 B3M0D8 B4JV86 B3NZU1 Q9VHS8 A0A0K8UBI3 A0A034VAP8 A0A1B6C2G1 A0A0L0CBE0 B4HLL9 A0A3B0JQK9 T1PG03 Q298G7 B4G3R2 A0A1I8Q8H2 A0A1A9WML1 A0A1B0FFV4 A0A1A9YQ17 A0A1A9UFH7 A0A1B0B4Y7 A0A1A9ZT23 E0W2V0 A0A3S3SQE4 A0A1S3J3P5 T1K100 A0A2S1TJI0 A0A0K2T8I3 G3MPX6 A0A023GJK7 A0A0P5URV6 A0A0P6BGL6 T1E1I3 L7M7P7 A0A131XB35 A0A0P5CLK1 A0A0P5VWE4 C1C2L4
I4DME1 I4DJR8 A0A194PFX7 A0A0N1PHX3 S4PWN2 A0A1Q3FV91 A0A2J7QNB4 A0A0M8ZSQ6 A0A067RMC6 A0A1B0CXD4 A0A154P503 A0A182J1G5 A0A2A3ERN3 A0A087ZZS7 A0A0L7R1R2 E2BEH0 A0A182QXS5 A0A182N6M7 A0A084W917 Q16GE2 A0A182VJT6 A0A1B0DM43 A0A182P301 A0A182MFM2 A0A182SNI4 A0A182UES5 A0A182X8D7 Q7PQQ2 A0A182RXB9 A0A182HIZ1 A0A1L8DGW1 A0A0J7KKY9 E9IZK2 A0A2M3Z6Y1 A0A2M4AH50 A0A2M4BQA4 W5JLN7 A0A151J4R6 A0A182XVP1 A0A151JVP6 A0A182JWR5 A0A026WX81 A0A195CSB2 A0A158NIN0 E2AWS1 A0A0K8TMU1 A0A1L8DGS9 A0A1B6L3M7 A0A232ELK5 W8BID8 A0A1B6FKM8 A0A1B6IUQ3 B4NGN0 A0A1W4VH90 B4LYN9 B4K8T2 B4PT42 A0A0M3QX77 B3M0D8 B4JV86 B3NZU1 Q9VHS8 A0A0K8UBI3 A0A034VAP8 A0A1B6C2G1 A0A0L0CBE0 B4HLL9 A0A3B0JQK9 T1PG03 Q298G7 B4G3R2 A0A1I8Q8H2 A0A1A9WML1 A0A1B0FFV4 A0A1A9YQ17 A0A1A9UFH7 A0A1B0B4Y7 A0A1A9ZT23 E0W2V0 A0A3S3SQE4 A0A1S3J3P5 T1K100 A0A2S1TJI0 A0A0K2T8I3 G3MPX6 A0A023GJK7 A0A0P5URV6 A0A0P6BGL6 T1E1I3 L7M7P7 A0A131XB35 A0A0P5CLK1 A0A0P5VWE4 C1C2L4
PDB
2J0S
E-value=0,
Score=1834
Ontologies
PATHWAY
GO
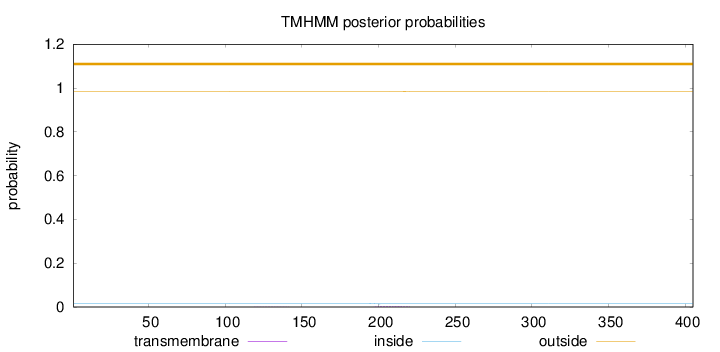
Topology
Subcellular location
Nucleus
Length:
405
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0630099999999999
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01725
outside
1 - 405
Population Genetic Test Statistics
Pi
254.378964
Theta
174.591154
Tajima's D
1.33245
CLR
0.031631
CSRT
0.752712364381781
Interpretation
Uncertain
