Gene
KWMTBOMO15052
Pre Gene Modal
BGIBMGA009860
Annotation
PREDICTED:_piggyBac_transposable_element-derived_protein_4-like_isoform_X4_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.08
Sequence
CDS
ATGGAATCTCAACGTGCTGCAGCAGGTCGAGGCAGAAGTGGTCGTGCTGGCGGTCAACGTGTTCGTGGTCATCACCAAGCCCGACCTTCGGCCTCTGAATCACTCGTCACTATGCCAATCGAGGCGTTGCCGGCGGGTCGAGCGATGGAGACGCCGATGCACCCGAAGAGGGTAAGGGTGCTCGAGCCCGTAGTGGAAAACCCTAGATCAGAAATGTGGGGATCCAACGCTTCGTCCCCAGCCAGGAGGGACCAGGAGACGCCTCCGGTGCCATCACCTGCCCTATACGTAGAATGTGGGCCTAGCAGCGGGGCAAGCACGCCGACCACCGAGCGGCCTGCAAGTCTGAGTCACGTCTCAAAGGAAGCTCGGCGTGAATTAATCCCTTCGTCCTCTTCGTCCACTCGACCTCTTCGCGACGAAGAGATTTTGACATTGCTCAATTACGGCAGCGAAGAAGAGTCGGAGGAAGAGGAGGACGAAGAGACTATGCAGCGGCTGACGGTGCCGCACTCGACCAGGATGTGGTTAGACGAAGAGGATGAGGAAGTTGAAAGAGGTGAAGAGAGGATCCCTCCTCTCCCGAGAGATAACATTAATATAGAGCAACCTCCTCAACCTCTTCGTCTATCCACATCCACCTCACCGACGGCTCTTTCGCCCTCTGTGGCGCTAGGGCCAACTAATAAATTTATGTTTGAATGGAGAACGGCGCCAATGCCTGCAATCGAACCTGATTTAAGGCGAGAACCGTTCTCCCAAATTAGTGGTCCAAAGGTTTCTTTCGCGACTCCGTATGATGCATTCATTGCCATATGGGATCGTGAAATAATGGAATCGATAGTCGAGGAAACCAACATTTACGCACAGCAGCTGGCAACGGTCATGCTGGAGACTGGGACGATCCGCCCTCATAGCTGGATTACTCGATGGCAAGATACGGATGTAAACGAACTTTACACGTATTTTGCTATCATTCTGGCTATGGGGGTGTTCATCAAAAGCTGCCTGACCGAGTATTGGTGCACGGCACAAGACGTATTTTATACACCAGGCTTCTCCGCCCAAATGTCGTATGACCGTTTCCGGCTGCTGTCAAGGTGCCTACACTTTTCTAATAACACTGCTTGTGATCTTGCAATGTTGACACGACGCCAGGTCATGCTGTACAAAATACAACCGTTAATAGACCACTTAAATAAAAAATTTGCGGAACTTTACAATTTGGGTCAAAATTTAGCAGTGGATGAAAGTTTGACGATGTGGAAAGGCTGGCTTGATATAAACCAGTTCATTCCACAAAAGGCAGCGACAGTTGGGATCAAAACTTATGAAGTTTGTGAATCGCAGAGTGGTTACCTGTGGCGGTTTGAAGTTCACGCTGGCCATGACATCTCGGCTTCACAGGACGATCCAATTTCTGGCACAGTACCAGCTCTCGTGCTCAGACTTTTAAATGGTCTTGAGCATAAAGGGCACACAATATGGATGGATAATTTTTATAACTCTCCAGCGCTAGCCCGAGAACTTAAGACACGCGGCTTTGACTGTGTGGGCACACTGCGGACTAACCGACAATTTGTGCCGTACGAAGTGACGTCCCTTACAAAAAGGGATATGACAGTCGGTCAAGTGATGGGATGCACCTCGGGAGATGTGGATATTTTGGTGTGGAAAGATAAAGGCCGGGTGGCATTTATTTCCACATACCATGGTCTTGCATCTGTTACGTTTGAGGAAAGACTGAAGCCGACTGTCGCGTACGATTATAATATTTGTATGGGCGGAGTAGACCGCAAGGACCAGCAGCTTGCAGTGTATCCCATAGAAAGAAGAAGGACAAAAGTTTGGTACAAGAGATTCTTTAGAAGGCTCTTGAACGCTAGTGTCTTGAACTCGCACATACTACACCAAAATCAGTCATTCTCACACCGGAAATTCCGGACGGTGTTAATCACTGAACTATTGAGTGCACACCGTACACAACCACCTAGCACACTCACCCCAACACATGTACATTGCCCGGCGCAGTATGGCCTGACCAAGAGCGGTAAATCAGACCGCCTTAGGCGCCAGTGTGCCGTCTGCTCCAGGCGGACTCACACATATTGCAAGGCCTGCAACGTGGCAATGTGCATTTTCACTTGCTACGAGACATACCACACTACACATCGACACTTTACCGCCACCTCAGTACCATGTGAGAGGATTTTTTCAAAATCTGGTAAACTCATTTCTGACCGACGTAACAGGCTTTCGGTAGAAAAAGTAAAAAAGCTGATGTTTTTATGCACCAATAAGAAATAA
Protein
MESQRAAAGRGRSGRAGGQRVRGHHQARPSASESLVTMPIEALPAGRAMETPMHPKRVRVLEPVVENPRSEMWGSNASSPARRDQETPPVPSPALYVECGPSSGASTPTTERPASLSHVSKEARRELIPSSSSSTRPLRDEEILTLLNYGSEEESEEEEDEETMQRLTVPHSTRMWLDEEDEEVERGEERIPPLPRDNINIEQPPQPLRLSTSTSPTALSPSVALGPTNKFMFEWRTAPMPAIEPDLRREPFSQISGPKVSFATPYDAFIAIWDREIMESIVEETNIYAQQLATVMLETGTIRPHSWITRWQDTDVNELYTYFAIILAMGVFIKSCLTEYWCTAQDVFYTPGFSAQMSYDRFRLLSRCLHFSNNTACDLAMLTRRQVMLYKIQPLIDHLNKKFAELYNLGQNLAVDESLTMWKGWLDINQFIPQKAATVGIKTYEVCESQSGYLWRFEVHAGHDISASQDDPISGTVPALVLRLLNGLEHKGHTIWMDNFYNSPALARELKTRGFDCVGTLRTNRQFVPYEVTSLTKRDMTVGQVMGCTSGDVDILVWKDKGRVAFISTYHGLASVTFEERLKPTVAYDYNICMGGVDRKDQQLAVYPIERRRTKVWYKRFFRRLLNASVLNSHILHQNQSFSHRKFRTVLITELLSAHRTQPPSTLTPTHVHCPAQYGLTKSGKSDRLRRQCAVCSRRTHTYCKACNVAMCIFTCYETYHTTHRHFTATSVPCERIFSKSGKLISDRRNRLSVEKVKKLMFLCTNKK
Summary
Uniprot
Pubmed
EMBL
Proteomes
Pfam
PF13843 DDE_Tnp_1_7
Interpro
IPR029526
PGBD
ProteinModelPortal
Ontologies
GO
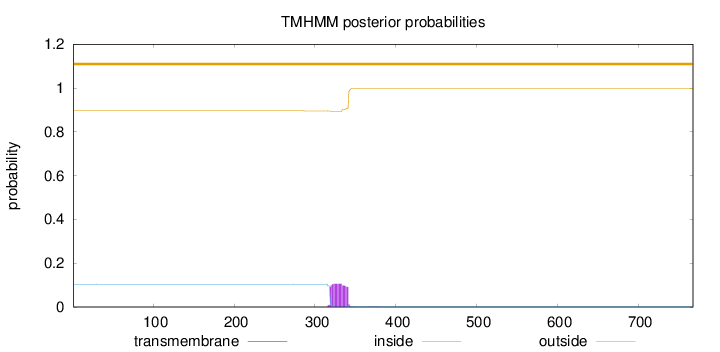
Topology
Length:
768
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.38524000000001
Exp number, first 60 AAs:
0.00079
Total prob of N-in:
0.10427
outside
1 - 768
Population Genetic Test Statistics
Pi
196.004866
Theta
190.574965
Tajima's D
1.431618
CLR
0.560114
CSRT
0.773961301934903
Interpretation
Uncertain
