Gene
KWMTBOMO15051 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA004857
Annotation
PREDICTED:_adenylosuccinate_synthetase-like_[Bombyx_mori]
Full name
Adenylosuccinate synthetase
+ More
Adenylosuccinate synthetase isozyme 1
Adenylosuccinate synthetase isozyme 1
Alternative Name
IMP--aspartate ligase
Adenylosuccinate synthetase, basic isozyme
Adenylosuccinate synthetase, muscle isozyme
IMP--aspartate ligase 1
Adenylosuccinate synthetase, basic isozyme
Adenylosuccinate synthetase, muscle isozyme
IMP--aspartate ligase 1
Location in the cell
Cytoplasmic Reliability : 2.327
Sequence
CDS
ATGGTTACAACAGTGAATTCGATTAATACAGAAATTAACGGTGAATTGTCGCGTTCGTGCCCATCAAAGAAAACCAAAATGGAAAACAAAGTAACTGTTGTTTTGGGTGCTCAGTGGGGCGATGAGGGCAAAGGAAAAGTTGTAGATTTACTTGCGGTGGATTCCGACATTGTTTGCCGTTGTCAGGGCGGGAACAACGCTGGACACACAGTGGTAGTTGACGGCAAAGAATTTGACTTCCATCTGTTGCCAAGTGGCATCATCAACCAGAAGTGCACGTCTGTCATAGGTAATGGCGTGGTGATCCACCTGCCCGGACTGTTTGAGGAGCTGAAGAAGAACGAGCTGAAAGGCATGAAGGGTTGCGAAGGCAGACTCGTGATCTCGGACAGGGCTCATCTTGTTTTCGATATACATCAACAGGTATAG
Protein
MVTTVNSINTEINGELSRSCPSKKTKMENKVTVVLGAQWGDEGKGKVVDLLAVDSDIVCRCQGGNNAGHTVVVDGKEFDFHLLPSGIINQKCTSVIGNGVVIHLPGLFEELKKNELKGMKGCEGRLVISDRAHLVFDIHQQV
Summary
Description
Plays an important role in the de novo pathway and in the salvage pathway of purine nucleotide biosynthesis. Catalyzes the first commited step in the biosynthesis of AMP from IMP.
Plays an important role in the de novo pathway of purine nucleotide biosynthesis.
Component of the purine nucleotide cycle (PNC), which interconverts IMP and AMP to regulate the nucleotide levels in various tissues, and which contributes to glycolysis and ammoniagenesis. Catalyzes the first commited step in the biosynthesis of AMP from IMP.
Plays an important role in the de novo pathway and in the salvage pathway of purine nucleotide biosynthesis. Catalyzes the first committed step in the biosynthesis of AMP from IMP (By similarity).
Component of the purine nucleotide cycle (PNC), which interconverts IMP and AMP to regulate the nucleotide levels in various tissues, and which contributes to glycolysis and ammoniagenesis. Catalyzes the first committed step in the biosynthesis of AMP from IMP.
Plays an important role in the de novo pathway of purine nucleotide biosynthesis.
Component of the purine nucleotide cycle (PNC), which interconverts IMP and AMP to regulate the nucleotide levels in various tissues, and which contributes to glycolysis and ammoniagenesis. Catalyzes the first commited step in the biosynthesis of AMP from IMP.
Plays an important role in the de novo pathway and in the salvage pathway of purine nucleotide biosynthesis. Catalyzes the first committed step in the biosynthesis of AMP from IMP (By similarity).
Component of the purine nucleotide cycle (PNC), which interconverts IMP and AMP to regulate the nucleotide levels in various tissues, and which contributes to glycolysis and ammoniagenesis. Catalyzes the first committed step in the biosynthesis of AMP from IMP.
Catalytic Activity
GTP + IMP + L-aspartate = GDP + 2 H(+) + N(6)-(1,2-dicarboxyethyl)-AMP + phosphate
Cofactor
Mg(2+)
Subunit
Homodimer.
Similarity
Belongs to the adenylosuccinate synthetase family.
Keywords
Complete proteome
Cytoplasm
GTP-binding
Ligase
Magnesium
Metal-binding
Nucleotide-binding
Purine biosynthesis
Reference proteome
Phosphoprotein
Feature
chain Adenylosuccinate synthetase
Uniprot
A0A2H1W6Y8
A0A220K8N8
A0A3S2PKC8
A0A194PGM4
S4PBN4
A0A212ETE7
+ More
V9ICX8 E2BWT4 A0A026WH21 A0A0J7KG59 A0A087ZXT6 A0A2A3EAG2 E2A7N5 A0A1B6DNJ2 A0A154PDF2 A0A195CK02 A0A2D0RLL9 A0A3Q1GDU1 A0A2P8XV56 A0A3B4WWZ8 A0A3Q1EYT2 A0A067QMR7 A0A3Q0S1J8 A0A151I0Q4 A0A151X5C1 A0A158NAF0 F4WMJ2 A0A182PS43 E9I9P3 A0A195EVK5 A0A2J7R6L9 A0A151J0F6 A0A2M4CV48 A0A1A8V891 A0A2M4CUQ0 H2ZYA8 A0A2M4AA55 A0A2M4BML8 A0A0S7GLR8 A0A2D4L0B1 A0A3B3RGB3 A0A3Q2QGT3 A0A2S2NLH3 Q17G75 A0A3Q3AWN5 A0A060WG10 A0A2M4CUS8 A0A1A7WDA1 A0A182FA55 A0A3B4CTF0 A0A3N0XLU3 A0A3Q3GN41 A0A3Q1HML1 A0A182GDW5 A0A023ETL6 A0A2I4BI08 G3NSJ9 A0A3Q1F0S9 A0A3B1K2H3 F6TXD9 A0A3Q0S1A6 T1D2C0 A0A3Q1GBA2 A0A2M3Z1E3 A0A336LYM2 A0A3Q0J6D1 A0A3Q2P5T9 A0A182JBY9 A0A182NKU2 V9KKL2 W5KYJ9 A0A182R802 A0A0P7VU47 T1D2A9 A0A336KE12 A0A1W4Y2Z2 A0A3S2P4I4 U5EUC9 A0A060VPG7 A0A2M3ZG43 A0A3P9IBG6 B9EL12 A0A084VZC5 A0A1Y1JWT5 A0A3P9K396 H2MR87 A0A2M3Z1D7 Q6NSN5 A0A0K8TT85 A0A3Q0S720 A0A0C9Q2C0 A0A3Q0S6F6 I3K2F0 A0A2R7WE21 H3DJA4 Q4RLN9 A0A1S6KZC4 A0A3P8UCN4 A0A3Q1B364 A0A3P8RK35
V9ICX8 E2BWT4 A0A026WH21 A0A0J7KG59 A0A087ZXT6 A0A2A3EAG2 E2A7N5 A0A1B6DNJ2 A0A154PDF2 A0A195CK02 A0A2D0RLL9 A0A3Q1GDU1 A0A2P8XV56 A0A3B4WWZ8 A0A3Q1EYT2 A0A067QMR7 A0A3Q0S1J8 A0A151I0Q4 A0A151X5C1 A0A158NAF0 F4WMJ2 A0A182PS43 E9I9P3 A0A195EVK5 A0A2J7R6L9 A0A151J0F6 A0A2M4CV48 A0A1A8V891 A0A2M4CUQ0 H2ZYA8 A0A2M4AA55 A0A2M4BML8 A0A0S7GLR8 A0A2D4L0B1 A0A3B3RGB3 A0A3Q2QGT3 A0A2S2NLH3 Q17G75 A0A3Q3AWN5 A0A060WG10 A0A2M4CUS8 A0A1A7WDA1 A0A182FA55 A0A3B4CTF0 A0A3N0XLU3 A0A3Q3GN41 A0A3Q1HML1 A0A182GDW5 A0A023ETL6 A0A2I4BI08 G3NSJ9 A0A3Q1F0S9 A0A3B1K2H3 F6TXD9 A0A3Q0S1A6 T1D2C0 A0A3Q1GBA2 A0A2M3Z1E3 A0A336LYM2 A0A3Q0J6D1 A0A3Q2P5T9 A0A182JBY9 A0A182NKU2 V9KKL2 W5KYJ9 A0A182R802 A0A0P7VU47 T1D2A9 A0A336KE12 A0A1W4Y2Z2 A0A3S2P4I4 U5EUC9 A0A060VPG7 A0A2M3ZG43 A0A3P9IBG6 B9EL12 A0A084VZC5 A0A1Y1JWT5 A0A3P9K396 H2MR87 A0A2M3Z1D7 Q6NSN5 A0A0K8TT85 A0A3Q0S720 A0A0C9Q2C0 A0A3Q0S6F6 I3K2F0 A0A2R7WE21 H3DJA4 Q4RLN9 A0A1S6KZC4 A0A3P8UCN4 A0A3Q1B364 A0A3P8RK35
EC Number
6.3.4.4
Pubmed
28630352
26354079
23622113
22118469
20798317
24508170
+ More
30249741 29403074 24845553 21347285 21719571 21282665 9215903 29240929 17510324 24755649 26483478 24945155 25329095 17495919 24330624 24402279 17554307 20433749 24438588 28004739 18307296 26506222 26369729 25186727 15496914 24330026 24487278
30249741 29403074 24845553 21347285 21719571 21282665 9215903 29240929 17510324 24755649 26483478 24945155 25329095 17495919 24330624 24402279 17554307 20433749 24438588 28004739 18307296 26506222 26369729 25186727 15496914 24330026 24487278
EMBL
ODYU01006731
SOQ48840.1
MF319738
ASJ26452.1
RSAL01000008
RVE53928.1
+ More
KQ459604 KPI92183.1 GAIX01005822 JAA86738.1 AGBW02012595 OWR44749.1 JR038558 AEY58316.1 GL451188 EFN79798.1 KK107231 QOIP01000009 EZA54971.1 RLU18603.1 LBMM01008054 KMQ89186.1 KZ288308 PBC28708.1 GL437365 EFN70556.1 GEDC01020859 GEDC01010052 JAS16439.1 JAS27246.1 KQ434869 KZC09288.1 KQ977642 KYN01043.1 PYGN01001299 PSN35890.1 KK853153 KDR10573.1 KQ976600 KYM79472.1 KQ982519 KYQ55489.1 ADTU01010272 GL888217 EGI64603.1 GL761846 EFZ22633.1 KQ981954 KYN32176.1 NEVH01006756 PNF36482.1 KQ980629 KYN14996.1 GGFL01004893 MBW69071.1 HAEJ01015325 SBS55782.1 GGFL01004894 MBW69072.1 AFYH01002014 AFYH01002015 AFYH01002016 AFYH01002017 AFYH01002018 AFYH01002019 AFYH01002020 AFYH01002021 GGFK01004301 MBW37622.1 GGFJ01005179 MBW54320.1 GBYX01451398 JAO30108.1 IACL01112146 LAB14404.1 GGMR01005414 MBY18033.1 CH477265 FR904523 CDQ65951.1 GGFL01004895 MBW69073.1 HADW01002305 SBP03705.1 RJVU01069573 ROI68631.1 JXUM01056782 JXUM01056783 JXUM01056784 KQ561925 KXJ77138.1 GAPW01001382 JAC12216.1 GALA01001673 JAA93179.1 GGFM01001582 MBW22333.1 UFQS01000309 UFQT01000309 SSX02708.1 SSX23082.1 JW866046 AFO98563.1 JARO02000699 KPP77688.1 GALA01001688 JAA93164.1 SSX02707.1 SSX23081.1 CM012458 RVE57099.1 GANO01001537 JAB58334.1 FR904269 CDQ56828.1 GGFM01006751 MBW27502.1 BT056337 ACM08209.1 ATLV01018738 ATLV01018739 ATLV01018740 KE525248 KFB43319.1 GEZM01098728 JAV53784.1 GGFM01001591 MBW22342.1 BC070009 GDAI01000242 JAI17361.1 GBYB01008148 JAG77915.1 AERX01018666 KK854669 PTY17759.1 CAAE01015019 CAG10693.1 KC223176 AQT27079.1
KQ459604 KPI92183.1 GAIX01005822 JAA86738.1 AGBW02012595 OWR44749.1 JR038558 AEY58316.1 GL451188 EFN79798.1 KK107231 QOIP01000009 EZA54971.1 RLU18603.1 LBMM01008054 KMQ89186.1 KZ288308 PBC28708.1 GL437365 EFN70556.1 GEDC01020859 GEDC01010052 JAS16439.1 JAS27246.1 KQ434869 KZC09288.1 KQ977642 KYN01043.1 PYGN01001299 PSN35890.1 KK853153 KDR10573.1 KQ976600 KYM79472.1 KQ982519 KYQ55489.1 ADTU01010272 GL888217 EGI64603.1 GL761846 EFZ22633.1 KQ981954 KYN32176.1 NEVH01006756 PNF36482.1 KQ980629 KYN14996.1 GGFL01004893 MBW69071.1 HAEJ01015325 SBS55782.1 GGFL01004894 MBW69072.1 AFYH01002014 AFYH01002015 AFYH01002016 AFYH01002017 AFYH01002018 AFYH01002019 AFYH01002020 AFYH01002021 GGFK01004301 MBW37622.1 GGFJ01005179 MBW54320.1 GBYX01451398 JAO30108.1 IACL01112146 LAB14404.1 GGMR01005414 MBY18033.1 CH477265 FR904523 CDQ65951.1 GGFL01004895 MBW69073.1 HADW01002305 SBP03705.1 RJVU01069573 ROI68631.1 JXUM01056782 JXUM01056783 JXUM01056784 KQ561925 KXJ77138.1 GAPW01001382 JAC12216.1 GALA01001673 JAA93179.1 GGFM01001582 MBW22333.1 UFQS01000309 UFQT01000309 SSX02708.1 SSX23082.1 JW866046 AFO98563.1 JARO02000699 KPP77688.1 GALA01001688 JAA93164.1 SSX02707.1 SSX23081.1 CM012458 RVE57099.1 GANO01001537 JAB58334.1 FR904269 CDQ56828.1 GGFM01006751 MBW27502.1 BT056337 ACM08209.1 ATLV01018738 ATLV01018739 ATLV01018740 KE525248 KFB43319.1 GEZM01098728 JAV53784.1 GGFM01001591 MBW22342.1 BC070009 GDAI01000242 JAI17361.1 GBYB01008148 JAG77915.1 AERX01018666 KK854669 PTY17759.1 CAAE01015019 CAG10693.1 KC223176 AQT27079.1
Proteomes
UP000283053
UP000053268
UP000007151
UP000008237
UP000053097
UP000279307
+ More
UP000036403 UP000005203 UP000242457 UP000000311 UP000076502 UP000078542 UP000221080 UP000257200 UP000245037 UP000261360 UP000027135 UP000261340 UP000078540 UP000075809 UP000005205 UP000007755 UP000075885 UP000078541 UP000235965 UP000078492 UP000008672 UP000261540 UP000265000 UP000008820 UP000264800 UP000193380 UP000069272 UP000261440 UP000261660 UP000265040 UP000069940 UP000249989 UP000192220 UP000007635 UP000018467 UP000002280 UP000079169 UP000075880 UP000075884 UP000075900 UP000034805 UP000192224 UP000265200 UP000030765 UP000265180 UP000001038 UP000000437 UP000005207 UP000007303 UP000265120 UP000257160 UP000265080
UP000036403 UP000005203 UP000242457 UP000000311 UP000076502 UP000078542 UP000221080 UP000257200 UP000245037 UP000261360 UP000027135 UP000261340 UP000078540 UP000075809 UP000005205 UP000007755 UP000075885 UP000078541 UP000235965 UP000078492 UP000008672 UP000261540 UP000265000 UP000008820 UP000264800 UP000193380 UP000069272 UP000261440 UP000261660 UP000265040 UP000069940 UP000249989 UP000192220 UP000007635 UP000018467 UP000002280 UP000079169 UP000075880 UP000075884 UP000075900 UP000034805 UP000192224 UP000265200 UP000030765 UP000265180 UP000001038 UP000000437 UP000005207 UP000007303 UP000265120 UP000257160 UP000265080
PRIDE
Pfam
PF00709 Adenylsucc_synt
Interpro
SUPFAM
SSF52540
SSF52540
Gene 3D
CDD
ProteinModelPortal
A0A2H1W6Y8
A0A220K8N8
A0A3S2PKC8
A0A194PGM4
S4PBN4
A0A212ETE7
+ More
V9ICX8 E2BWT4 A0A026WH21 A0A0J7KG59 A0A087ZXT6 A0A2A3EAG2 E2A7N5 A0A1B6DNJ2 A0A154PDF2 A0A195CK02 A0A2D0RLL9 A0A3Q1GDU1 A0A2P8XV56 A0A3B4WWZ8 A0A3Q1EYT2 A0A067QMR7 A0A3Q0S1J8 A0A151I0Q4 A0A151X5C1 A0A158NAF0 F4WMJ2 A0A182PS43 E9I9P3 A0A195EVK5 A0A2J7R6L9 A0A151J0F6 A0A2M4CV48 A0A1A8V891 A0A2M4CUQ0 H2ZYA8 A0A2M4AA55 A0A2M4BML8 A0A0S7GLR8 A0A2D4L0B1 A0A3B3RGB3 A0A3Q2QGT3 A0A2S2NLH3 Q17G75 A0A3Q3AWN5 A0A060WG10 A0A2M4CUS8 A0A1A7WDA1 A0A182FA55 A0A3B4CTF0 A0A3N0XLU3 A0A3Q3GN41 A0A3Q1HML1 A0A182GDW5 A0A023ETL6 A0A2I4BI08 G3NSJ9 A0A3Q1F0S9 A0A3B1K2H3 F6TXD9 A0A3Q0S1A6 T1D2C0 A0A3Q1GBA2 A0A2M3Z1E3 A0A336LYM2 A0A3Q0J6D1 A0A3Q2P5T9 A0A182JBY9 A0A182NKU2 V9KKL2 W5KYJ9 A0A182R802 A0A0P7VU47 T1D2A9 A0A336KE12 A0A1W4Y2Z2 A0A3S2P4I4 U5EUC9 A0A060VPG7 A0A2M3ZG43 A0A3P9IBG6 B9EL12 A0A084VZC5 A0A1Y1JWT5 A0A3P9K396 H2MR87 A0A2M3Z1D7 Q6NSN5 A0A0K8TT85 A0A3Q0S720 A0A0C9Q2C0 A0A3Q0S6F6 I3K2F0 A0A2R7WE21 H3DJA4 Q4RLN9 A0A1S6KZC4 A0A3P8UCN4 A0A3Q1B364 A0A3P8RK35
V9ICX8 E2BWT4 A0A026WH21 A0A0J7KG59 A0A087ZXT6 A0A2A3EAG2 E2A7N5 A0A1B6DNJ2 A0A154PDF2 A0A195CK02 A0A2D0RLL9 A0A3Q1GDU1 A0A2P8XV56 A0A3B4WWZ8 A0A3Q1EYT2 A0A067QMR7 A0A3Q0S1J8 A0A151I0Q4 A0A151X5C1 A0A158NAF0 F4WMJ2 A0A182PS43 E9I9P3 A0A195EVK5 A0A2J7R6L9 A0A151J0F6 A0A2M4CV48 A0A1A8V891 A0A2M4CUQ0 H2ZYA8 A0A2M4AA55 A0A2M4BML8 A0A0S7GLR8 A0A2D4L0B1 A0A3B3RGB3 A0A3Q2QGT3 A0A2S2NLH3 Q17G75 A0A3Q3AWN5 A0A060WG10 A0A2M4CUS8 A0A1A7WDA1 A0A182FA55 A0A3B4CTF0 A0A3N0XLU3 A0A3Q3GN41 A0A3Q1HML1 A0A182GDW5 A0A023ETL6 A0A2I4BI08 G3NSJ9 A0A3Q1F0S9 A0A3B1K2H3 F6TXD9 A0A3Q0S1A6 T1D2C0 A0A3Q1GBA2 A0A2M3Z1E3 A0A336LYM2 A0A3Q0J6D1 A0A3Q2P5T9 A0A182JBY9 A0A182NKU2 V9KKL2 W5KYJ9 A0A182R802 A0A0P7VU47 T1D2A9 A0A336KE12 A0A1W4Y2Z2 A0A3S2P4I4 U5EUC9 A0A060VPG7 A0A2M3ZG43 A0A3P9IBG6 B9EL12 A0A084VZC5 A0A1Y1JWT5 A0A3P9K396 H2MR87 A0A2M3Z1D7 Q6NSN5 A0A0K8TT85 A0A3Q0S720 A0A0C9Q2C0 A0A3Q0S6F6 I3K2F0 A0A2R7WE21 H3DJA4 Q4RLN9 A0A1S6KZC4 A0A3P8UCN4 A0A3Q1B364 A0A3P8RK35
PDB
2DGN
E-value=2.72852e-51,
Score=504
Ontologies
KEGG
PATHWAY
GO
PANTHER
Topology
Subcellular location
Cytoplasm
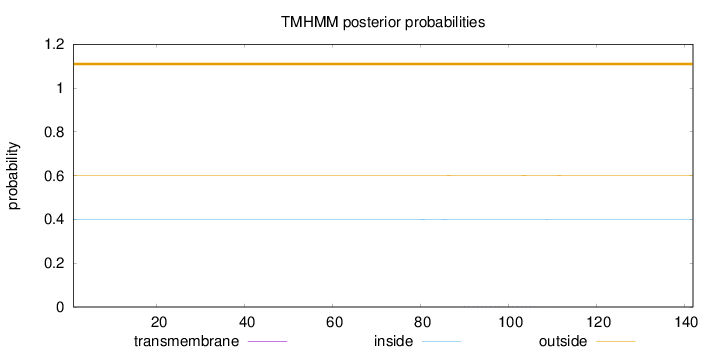
Length:
142
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.03612
Exp number, first 60 AAs:
0
Total prob of N-in:
0.39919
outside
1 - 142
Population Genetic Test Statistics
Pi
222.188762
Theta
159.154098
Tajima's D
1.880068
CLR
0.359958
CSRT
0.860806959652017
Interpretation
Uncertain
